
Aequorivita sublithincola (strain DSM 14238 / LMG 21431 / ACAM 643 / 9-3)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Aequorivita; Aequorivita sublithincola
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
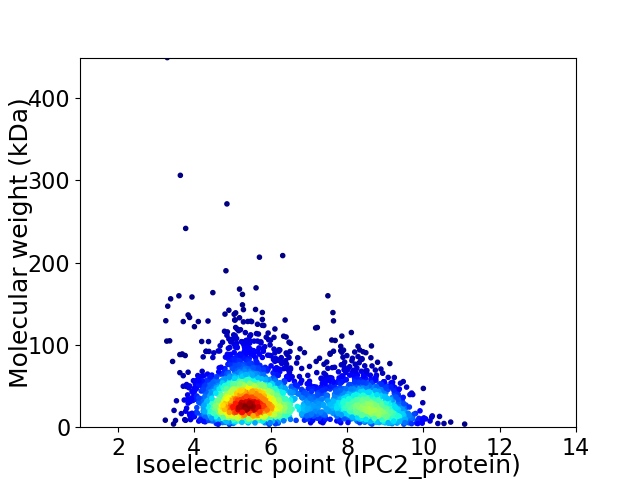
Virtual 2D-PAGE plot for 3134 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3Z094|I3Z094_AEQSU 4-hydroxy-tetrahydrodipicolinate synthase OS=Aequorivita sublithincola (strain DSM 14238 / LMG 21431 / ACAM 643 / 9-3) OX=746697 GN=dapA PE=3 SV=1
MM1 pKa = 7.63KK2 pKa = 10.37KK3 pKa = 8.4ITLLLVCLIGLSGMVAAQNVQSSSTVDD30 pKa = 3.3DD31 pKa = 4.04LLNRR35 pKa = 11.84LSQIGTNAGDD45 pKa = 3.91LSAYY49 pKa = 7.47FTKK52 pKa = 10.52QEE54 pKa = 3.81QQTLHH59 pKa = 6.65NYY61 pKa = 9.55FNLNKK66 pKa = 10.0NAPTSATVKK75 pKa = 9.8YY76 pKa = 9.56AKK78 pKa = 9.9NVNGSGFGDD87 pKa = 5.08FILTGNTQAIQLNYY101 pKa = 9.95KK102 pKa = 9.09VASDD106 pKa = 3.81VEE108 pKa = 4.77SNRR111 pKa = 11.84GVMAIIAYY119 pKa = 7.22TDD121 pKa = 3.24RR122 pKa = 11.84PTFEE126 pKa = 3.87GAYY129 pKa = 9.5TGTLVNEE136 pKa = 4.72DD137 pKa = 3.74FSGGPGAGSILACGPVMSSGGDD159 pKa = 3.21GCFAAGEE166 pKa = 4.36LEE168 pKa = 4.62DD169 pKa = 4.75GFSITASSGGDD180 pKa = 3.34TIYY183 pKa = 10.45IGAGAIGNTSTLVGANTFADD203 pKa = 3.72TTVLNFSPDD212 pKa = 2.82GAYY215 pKa = 10.31AVGMDD220 pKa = 4.51LFVDD224 pKa = 3.95SVGNADD230 pKa = 3.1IRR232 pKa = 11.84VYY234 pKa = 11.73DD235 pKa = 3.5MGGTLMDD242 pKa = 4.11TFTVSNTANTEE253 pKa = 3.83NFIGLISDD261 pKa = 3.76DD262 pKa = 4.83AIGKK266 pKa = 9.26IEE268 pKa = 4.01IQAEE272 pKa = 3.9ADD274 pKa = 3.22AGEE277 pKa = 4.35LFGNLAFGTDD287 pKa = 4.18PIGGGGSGPAVCFGANNTTSSIITFDD313 pKa = 3.86PADD316 pKa = 3.76PAAFTTLGTSPAPVFEE332 pKa = 4.37NAGAVDD338 pKa = 4.28PNDD341 pKa = 3.35DD342 pKa = 3.29TTAYY346 pKa = 10.56VLDD349 pKa = 4.18SGGLFYY355 pKa = 11.1SVDD358 pKa = 3.95LTTGVYY364 pKa = 9.75TNLGTILAPGGNQWSGAEE382 pKa = 4.15FDD384 pKa = 5.19PISGTLYY391 pKa = 10.43AISVNGALTATTLSTIDD408 pKa = 3.6IGALTATTIGLTGMAGGISLMIDD431 pKa = 3.26ANGDD435 pKa = 3.79GYY437 pKa = 11.62SHH439 pKa = 8.09DD440 pKa = 3.61IADD443 pKa = 3.83DD444 pKa = 3.26NFYY447 pKa = 11.22YY448 pKa = 10.81VDD450 pKa = 4.33LASGTASPIGPLGFDD465 pKa = 3.53ANFGQGGTWIDD476 pKa = 3.56GDD478 pKa = 3.92PGFVYY483 pKa = 10.67LSAFDD488 pKa = 4.01SGAFQSQWRR497 pKa = 11.84RR498 pKa = 11.84VDD500 pKa = 3.39VLTGSSTVIGLFNGGADD517 pKa = 3.52QVGWSSAKK525 pKa = 10.14GSLAVGIAEE534 pKa = 3.98NALEE538 pKa = 4.39GFSYY542 pKa = 10.95APNPTSGVLSLKK554 pKa = 10.45SINNIDD560 pKa = 3.28TVAIYY565 pKa = 11.39NMLGQNVMSSKK576 pKa = 10.32IGATTSDD583 pKa = 4.14LDD585 pKa = 3.55ISSLKK590 pKa = 9.07TGTYY594 pKa = 9.86IMQVTVAGQTAAFRR608 pKa = 11.84VLKK611 pKa = 10.69NN612 pKa = 3.33
MM1 pKa = 7.63KK2 pKa = 10.37KK3 pKa = 8.4ITLLLVCLIGLSGMVAAQNVQSSSTVDD30 pKa = 3.3DD31 pKa = 4.04LLNRR35 pKa = 11.84LSQIGTNAGDD45 pKa = 3.91LSAYY49 pKa = 7.47FTKK52 pKa = 10.52QEE54 pKa = 3.81QQTLHH59 pKa = 6.65NYY61 pKa = 9.55FNLNKK66 pKa = 10.0NAPTSATVKK75 pKa = 9.8YY76 pKa = 9.56AKK78 pKa = 9.9NVNGSGFGDD87 pKa = 5.08FILTGNTQAIQLNYY101 pKa = 9.95KK102 pKa = 9.09VASDD106 pKa = 3.81VEE108 pKa = 4.77SNRR111 pKa = 11.84GVMAIIAYY119 pKa = 7.22TDD121 pKa = 3.24RR122 pKa = 11.84PTFEE126 pKa = 3.87GAYY129 pKa = 9.5TGTLVNEE136 pKa = 4.72DD137 pKa = 3.74FSGGPGAGSILACGPVMSSGGDD159 pKa = 3.21GCFAAGEE166 pKa = 4.36LEE168 pKa = 4.62DD169 pKa = 4.75GFSITASSGGDD180 pKa = 3.34TIYY183 pKa = 10.45IGAGAIGNTSTLVGANTFADD203 pKa = 3.72TTVLNFSPDD212 pKa = 2.82GAYY215 pKa = 10.31AVGMDD220 pKa = 4.51LFVDD224 pKa = 3.95SVGNADD230 pKa = 3.1IRR232 pKa = 11.84VYY234 pKa = 11.73DD235 pKa = 3.5MGGTLMDD242 pKa = 4.11TFTVSNTANTEE253 pKa = 3.83NFIGLISDD261 pKa = 3.76DD262 pKa = 4.83AIGKK266 pKa = 9.26IEE268 pKa = 4.01IQAEE272 pKa = 3.9ADD274 pKa = 3.22AGEE277 pKa = 4.35LFGNLAFGTDD287 pKa = 4.18PIGGGGSGPAVCFGANNTTSSIITFDD313 pKa = 3.86PADD316 pKa = 3.76PAAFTTLGTSPAPVFEE332 pKa = 4.37NAGAVDD338 pKa = 4.28PNDD341 pKa = 3.35DD342 pKa = 3.29TTAYY346 pKa = 10.56VLDD349 pKa = 4.18SGGLFYY355 pKa = 11.1SVDD358 pKa = 3.95LTTGVYY364 pKa = 9.75TNLGTILAPGGNQWSGAEE382 pKa = 4.15FDD384 pKa = 5.19PISGTLYY391 pKa = 10.43AISVNGALTATTLSTIDD408 pKa = 3.6IGALTATTIGLTGMAGGISLMIDD431 pKa = 3.26ANGDD435 pKa = 3.79GYY437 pKa = 11.62SHH439 pKa = 8.09DD440 pKa = 3.61IADD443 pKa = 3.83DD444 pKa = 3.26NFYY447 pKa = 11.22YY448 pKa = 10.81VDD450 pKa = 4.33LASGTASPIGPLGFDD465 pKa = 3.53ANFGQGGTWIDD476 pKa = 3.56GDD478 pKa = 3.92PGFVYY483 pKa = 10.67LSAFDD488 pKa = 4.01SGAFQSQWRR497 pKa = 11.84RR498 pKa = 11.84VDD500 pKa = 3.39VLTGSSTVIGLFNGGADD517 pKa = 3.52QVGWSSAKK525 pKa = 10.14GSLAVGIAEE534 pKa = 3.98NALEE538 pKa = 4.39GFSYY542 pKa = 10.95APNPTSGVLSLKK554 pKa = 10.45SINNIDD560 pKa = 3.28TVAIYY565 pKa = 11.39NMLGQNVMSSKK576 pKa = 10.32IGATTSDD583 pKa = 4.14LDD585 pKa = 3.55ISSLKK590 pKa = 9.07TGTYY594 pKa = 9.86IMQVTVAGQTAAFRR608 pKa = 11.84VLKK611 pKa = 10.69NN612 pKa = 3.33
Molecular weight: 62.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3YV35|I3YV35_AEQSU ATP synthase subunit beta OS=Aequorivita sublithincola (strain DSM 14238 / LMG 21431 / ACAM 643 / 9-3) OX=746697 GN=atpD PE=3 SV=1
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.21KK16 pKa = 10.3RR17 pKa = 11.84KK18 pKa = 9.62KK19 pKa = 9.04RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.77KK27 pKa = 10.58KK28 pKa = 9.8KK29 pKa = 10.51KK30 pKa = 10.04
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.21KK16 pKa = 10.3RR17 pKa = 11.84KK18 pKa = 9.62KK19 pKa = 9.04RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.77KK27 pKa = 10.58KK28 pKa = 9.8KK29 pKa = 10.51KK30 pKa = 10.04
Molecular weight: 3.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1040270 |
29 |
4288 |
331.9 |
37.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.454 ± 0.042 | 0.724 ± 0.013 |
5.518 ± 0.042 | 6.905 ± 0.048 |
5.333 ± 0.04 | 6.522 ± 0.059 |
1.6 ± 0.021 | 7.896 ± 0.038 |
7.802 ± 0.062 | 9.222 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.254 ± 0.024 | 6.223 ± 0.047 |
3.455 ± 0.027 | 3.277 ± 0.025 |
3.35 ± 0.034 | 6.537 ± 0.035 |
5.791 ± 0.049 | 6.134 ± 0.036 |
1.012 ± 0.013 | 3.991 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
