
Sapporo virus (isolate GI/Human/Germany/pJG-Sap01) (Hu/Dresden/pJG-Sap01/DE)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Picornavirales; Caliciviridae; Sapovirus; Sapporo virus; Sapovirus isolates
Average proteome isoelectric point is 7.36
Get precalculated fractions of proteins
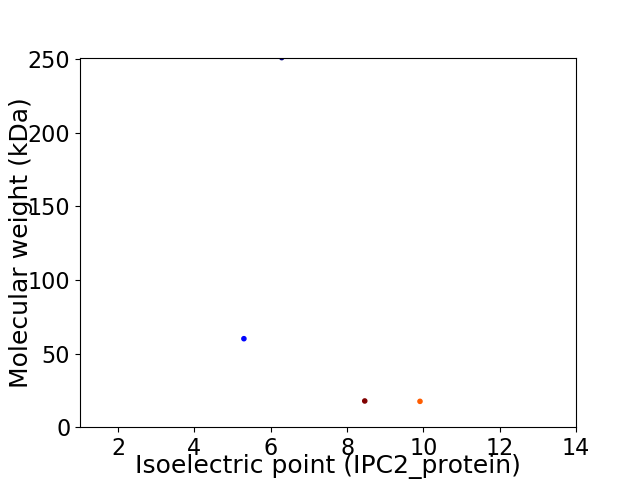
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q672I1-2|POLG-2_SVSAP Isoform of Q672I1 Isoform Subgenomic capsid protein of Genome polyprotein OS=Sapporo virus (isolate GI/Human/Germany/pJG-Sap01) OX=291175 PE=4 SV=1
MM1 pKa = 7.68EE2 pKa = 5.79GNGSNPEE9 pKa = 4.03PKK11 pKa = 9.91QSNNPMVVDD20 pKa = 4.25PPGTTGPTTSHH31 pKa = 5.31VVVANPEE38 pKa = 4.18QPNGAAQRR46 pKa = 11.84LEE48 pKa = 4.03LAVATGAIQSNVPEE62 pKa = 4.99AIRR65 pKa = 11.84NCFAVFRR72 pKa = 11.84TFAWNDD78 pKa = 3.16RR79 pKa = 11.84MPTGTFLGSISLHH92 pKa = 6.21PNINPYY98 pKa = 8.33TAHH101 pKa = 7.35LSGMWAGWGGSFEE114 pKa = 4.2VRR116 pKa = 11.84LSISGSGVFAGRR128 pKa = 11.84IIASVIPPGVDD139 pKa = 2.98PSSIRR144 pKa = 11.84DD145 pKa = 3.72PGVLPHH151 pKa = 6.89AFVDD155 pKa = 3.85ARR157 pKa = 11.84ITEE160 pKa = 4.41PVSFMIPDD168 pKa = 3.52VRR170 pKa = 11.84AVDD173 pKa = 3.6YY174 pKa = 11.25HH175 pKa = 8.42RR176 pKa = 11.84MDD178 pKa = 3.7GAEE181 pKa = 4.27PTCSLGFWVYY191 pKa = 10.8QPLLNPFSTTAVSTCWVSVEE211 pKa = 4.43TKK213 pKa = 10.3PGGDD217 pKa = 3.57FDD219 pKa = 4.81FCLLRR224 pKa = 11.84PPGQQMEE231 pKa = 4.51NGVSPEE237 pKa = 3.65GLLPRR242 pKa = 11.84RR243 pKa = 11.84LGYY246 pKa = 10.5SRR248 pKa = 11.84GNRR251 pKa = 11.84VGGLVVGMVLVAEE264 pKa = 4.88HH265 pKa = 6.14KK266 pKa = 10.11QVNRR270 pKa = 11.84HH271 pKa = 5.13FNSNSVTFGWSTAPVNPMAAEE292 pKa = 3.8IVTNQAHH299 pKa = 5.38STSRR303 pKa = 11.84HH304 pKa = 3.86AWLSIGAQNKK314 pKa = 9.37GPLFPGIPNHH324 pKa = 6.28FPDD327 pKa = 4.05SCASTIVGAMDD338 pKa = 3.72TSLGGRR344 pKa = 11.84PSTGVCGPAISFQNNGDD361 pKa = 3.5VYY363 pKa = 11.26EE364 pKa = 4.6NDD366 pKa = 3.7TPSVMFATYY375 pKa = 10.72DD376 pKa = 3.63PLTSGTGVALTNSINPASLALVRR399 pKa = 11.84ISNNDD404 pKa = 2.83FDD406 pKa = 4.46TSGFANDD413 pKa = 3.76KK414 pKa = 10.55NVVVQMSWEE423 pKa = 4.37MYY425 pKa = 9.52TGTNQIRR432 pKa = 11.84GQVTPMSGTNYY443 pKa = 9.37TFTSTGANTLVLWQEE458 pKa = 3.84RR459 pKa = 11.84MLSYY463 pKa = 10.78DD464 pKa = 3.05GHH466 pKa = 5.24QAILYY471 pKa = 9.11SSQLEE476 pKa = 3.96RR477 pKa = 11.84TAEE480 pKa = 3.91YY481 pKa = 9.83FQNDD485 pKa = 3.16IVNIPEE491 pKa = 3.5NSMAVFNVEE500 pKa = 3.98TNSASFQIGIRR511 pKa = 11.84PDD513 pKa = 3.24GYY515 pKa = 9.73MVTGGSIGINVPLEE529 pKa = 3.92PEE531 pKa = 3.84TRR533 pKa = 11.84FQYY536 pKa = 11.03VGILPLSAALSGPSGNMGRR555 pKa = 11.84ARR557 pKa = 11.84RR558 pKa = 11.84VFQQ561 pKa = 3.97
MM1 pKa = 7.68EE2 pKa = 5.79GNGSNPEE9 pKa = 4.03PKK11 pKa = 9.91QSNNPMVVDD20 pKa = 4.25PPGTTGPTTSHH31 pKa = 5.31VVVANPEE38 pKa = 4.18QPNGAAQRR46 pKa = 11.84LEE48 pKa = 4.03LAVATGAIQSNVPEE62 pKa = 4.99AIRR65 pKa = 11.84NCFAVFRR72 pKa = 11.84TFAWNDD78 pKa = 3.16RR79 pKa = 11.84MPTGTFLGSISLHH92 pKa = 6.21PNINPYY98 pKa = 8.33TAHH101 pKa = 7.35LSGMWAGWGGSFEE114 pKa = 4.2VRR116 pKa = 11.84LSISGSGVFAGRR128 pKa = 11.84IIASVIPPGVDD139 pKa = 2.98PSSIRR144 pKa = 11.84DD145 pKa = 3.72PGVLPHH151 pKa = 6.89AFVDD155 pKa = 3.85ARR157 pKa = 11.84ITEE160 pKa = 4.41PVSFMIPDD168 pKa = 3.52VRR170 pKa = 11.84AVDD173 pKa = 3.6YY174 pKa = 11.25HH175 pKa = 8.42RR176 pKa = 11.84MDD178 pKa = 3.7GAEE181 pKa = 4.27PTCSLGFWVYY191 pKa = 10.8QPLLNPFSTTAVSTCWVSVEE211 pKa = 4.43TKK213 pKa = 10.3PGGDD217 pKa = 3.57FDD219 pKa = 4.81FCLLRR224 pKa = 11.84PPGQQMEE231 pKa = 4.51NGVSPEE237 pKa = 3.65GLLPRR242 pKa = 11.84RR243 pKa = 11.84LGYY246 pKa = 10.5SRR248 pKa = 11.84GNRR251 pKa = 11.84VGGLVVGMVLVAEE264 pKa = 4.88HH265 pKa = 6.14KK266 pKa = 10.11QVNRR270 pKa = 11.84HH271 pKa = 5.13FNSNSVTFGWSTAPVNPMAAEE292 pKa = 3.8IVTNQAHH299 pKa = 5.38STSRR303 pKa = 11.84HH304 pKa = 3.86AWLSIGAQNKK314 pKa = 9.37GPLFPGIPNHH324 pKa = 6.28FPDD327 pKa = 4.05SCASTIVGAMDD338 pKa = 3.72TSLGGRR344 pKa = 11.84PSTGVCGPAISFQNNGDD361 pKa = 3.5VYY363 pKa = 11.26EE364 pKa = 4.6NDD366 pKa = 3.7TPSVMFATYY375 pKa = 10.72DD376 pKa = 3.63PLTSGTGVALTNSINPASLALVRR399 pKa = 11.84ISNNDD404 pKa = 2.83FDD406 pKa = 4.46TSGFANDD413 pKa = 3.76KK414 pKa = 10.55NVVVQMSWEE423 pKa = 4.37MYY425 pKa = 9.52TGTNQIRR432 pKa = 11.84GQVTPMSGTNYY443 pKa = 9.37TFTSTGANTLVLWQEE458 pKa = 3.84RR459 pKa = 11.84MLSYY463 pKa = 10.78DD464 pKa = 3.05GHH466 pKa = 5.24QAILYY471 pKa = 9.11SSQLEE476 pKa = 3.96RR477 pKa = 11.84TAEE480 pKa = 3.91YY481 pKa = 9.83FQNDD485 pKa = 3.16IVNIPEE491 pKa = 3.5NSMAVFNVEE500 pKa = 3.98TNSASFQIGIRR511 pKa = 11.84PDD513 pKa = 3.24GYY515 pKa = 9.73MVTGGSIGINVPLEE529 pKa = 3.92PEE531 pKa = 3.84TRR533 pKa = 11.84FQYY536 pKa = 11.03VGILPLSAALSGPSGNMGRR555 pKa = 11.84ARR557 pKa = 11.84RR558 pKa = 11.84VFQQ561 pKa = 3.97
Molecular weight: 60.11 kDa
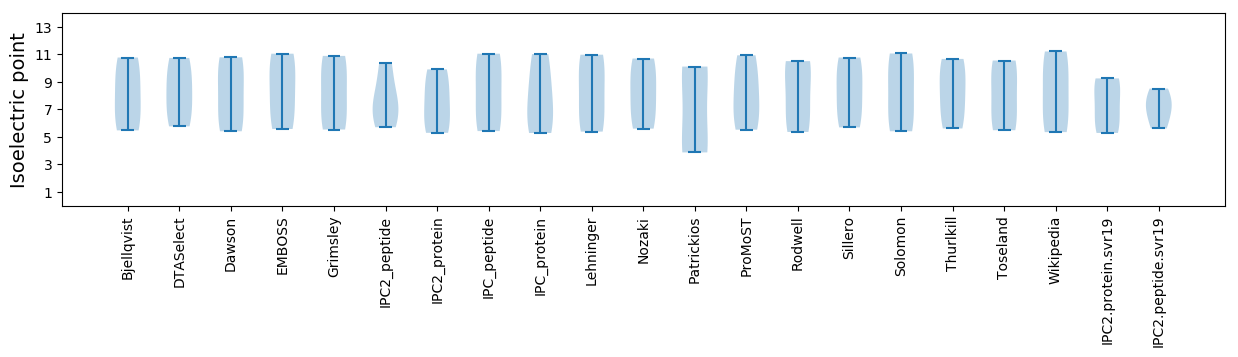
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q672I1|POLG_SVSAP Genome polyprotein OS=Sapporo virus (isolate GI/Human/Germany/pJG-Sap01) OX=291175 PE=3 SV=1
MM1 pKa = 7.85APTQSQSRR9 pKa = 11.84ATTRR13 pKa = 11.84WSLTRR18 pKa = 11.84LAQQVRR24 pKa = 11.84PHH26 pKa = 6.47PTLLLLIRR34 pKa = 11.84SNPMGPHH41 pKa = 6.1SAWSWLLPLVQSNPMSLRR59 pKa = 11.84QYY61 pKa = 9.5ATALQSFVLLLGTTGCPRR79 pKa = 11.84EE80 pKa = 4.46LFLDD84 pKa = 4.31LYY86 pKa = 11.01RR87 pKa = 11.84FIPTLTRR94 pKa = 11.84TLLTSLGCGPGGAVVLRR111 pKa = 11.84SGYY114 pKa = 9.93RR115 pKa = 11.84SLVLACSLGVSLLLSYY131 pKa = 10.59HH132 pKa = 6.22QGLIPRR138 pKa = 11.84PSGTQACCLTLSLMLASLSQFLSS161 pKa = 3.48
MM1 pKa = 7.85APTQSQSRR9 pKa = 11.84ATTRR13 pKa = 11.84WSLTRR18 pKa = 11.84LAQQVRR24 pKa = 11.84PHH26 pKa = 6.47PTLLLLIRR34 pKa = 11.84SNPMGPHH41 pKa = 6.1SAWSWLLPLVQSNPMSLRR59 pKa = 11.84QYY61 pKa = 9.5ATALQSFVLLLGTTGCPRR79 pKa = 11.84EE80 pKa = 4.46LFLDD84 pKa = 4.31LYY86 pKa = 11.01RR87 pKa = 11.84FIPTLTRR94 pKa = 11.84TLLTSLGCGPGGAVVLRR111 pKa = 11.84SGYY114 pKa = 9.93RR115 pKa = 11.84SLVLACSLGVSLLLSYY131 pKa = 10.59HH132 pKa = 6.22QGLIPRR138 pKa = 11.84PSGTQACCLTLSLMLASLSQFLSS161 pKa = 3.48
Molecular weight: 17.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3167 |
161 |
2280 |
791.8 |
86.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.241 ± 0.485 | 1.326 ± 0.306 |
4.168 ± 0.662 | 4.484 ± 0.759 |
4.2 ± 0.45 | 8.178 ± 0.585 |
2.4 ± 0.188 | 4.926 ± 0.55 |
2.873 ± 0.861 | 7.831 ± 2.349 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.716 ± 0.238 | 4.61 ± 0.877 |
6.599 ± 0.47 | 4.231 ± 0.729 |
5.526 ± 0.378 | 6.978 ± 1.492 |
8.304 ± 0.395 | 7.925 ± 0.868 |
1.674 ± 0.044 | 2.81 ± 0.455 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
