
Urechidicola croceus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Urechidicola
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
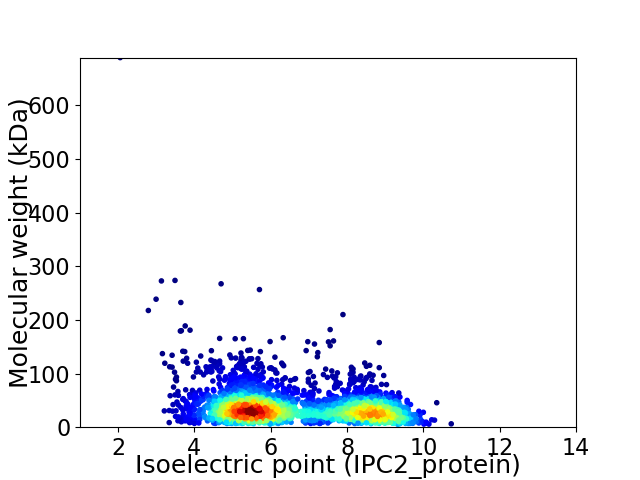
Virtual 2D-PAGE plot for 2946 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D8P9D9|A0A1D8P9D9_9FLAO Acyl-CoA thioesterase OS=Urechidicola croceus OX=1850246 GN=LPB138_11090 PE=3 SV=1
MM1 pKa = 6.91VFSVGFSQTVIYY13 pKa = 10.95SEE15 pKa = 4.92DD16 pKa = 3.57FEE18 pKa = 6.27SNIGTWTQDD27 pKa = 3.09TADD30 pKa = 3.74TLDD33 pKa = 2.92WSRR36 pKa = 11.84NSGGTGSGGTGPNNSSDD53 pKa = 3.12GSFYY57 pKa = 10.3MYY59 pKa = 10.59TEE61 pKa = 4.29ASSNYY66 pKa = 7.75NTTANFISEE75 pKa = 4.41PFNLSGISSPIFTFHH90 pKa = 5.98YY91 pKa = 10.82HH92 pKa = 5.16MFGSNMGTLNVNISTDD108 pKa = 3.24GGSTFGAPIWTQNGQHH124 pKa = 5.86QGSNSAAWQQVDD136 pKa = 3.81IDD138 pKa = 4.0LSTYY142 pKa = 10.17IGQTINIQFNGQVGSSYY159 pKa = 10.81RR160 pKa = 11.84SDD162 pKa = 3.0IAIDD166 pKa = 4.36EE167 pKa = 4.38ISLTVTSSPDD177 pKa = 3.28PEE179 pKa = 4.39IDD181 pKa = 3.39VTGNSLPIIGDD192 pKa = 3.48GTNAPSINDD201 pKa = 3.26NTDD204 pKa = 3.13FGTVQVGSEE213 pKa = 4.21TSIKK217 pKa = 10.06TYY219 pKa = 10.37TITNNGSLDD228 pKa = 3.8LVLGTSTISGSSDD241 pKa = 3.2FVISTFPGTTITPGNSEE258 pKa = 4.04IIEE261 pKa = 4.02ISFNTLSLGIQTAQLSIVNNDD282 pKa = 3.54PDD284 pKa = 4.02EE285 pKa = 4.43NPYY288 pKa = 10.63LINLSAEE295 pKa = 4.27GTQAFYY301 pKa = 11.09DD302 pKa = 3.71SDD304 pKa = 4.18GDD306 pKa = 4.43GVFDD310 pKa = 6.2DD311 pKa = 5.63VDD313 pKa = 3.9IDD315 pKa = 4.48DD316 pKa = 5.6DD317 pKa = 4.34NDD319 pKa = 4.71GIIDD323 pKa = 3.77SLEE326 pKa = 3.9EE327 pKa = 4.53SNCSGSIITNSVNYY341 pKa = 10.05KK342 pKa = 10.08FLNEE346 pKa = 4.05TFGTGGRR353 pKa = 11.84TTINTSYY360 pKa = 11.18NATTTYY366 pKa = 10.31CYY368 pKa = 10.68EE369 pKa = 5.06DD370 pKa = 3.7GTAGTDD376 pKa = 3.36TTGCPTLSDD385 pKa = 3.31TSLNDD390 pKa = 3.36GEE392 pKa = 4.5YY393 pKa = 9.72TVYY396 pKa = 10.99SRR398 pKa = 11.84ITNNDD403 pKa = 3.07DD404 pKa = 3.16VADD407 pKa = 5.25GINVDD412 pKa = 3.33IASWAEE418 pKa = 3.85DD419 pKa = 3.37FWYY422 pKa = 10.09EE423 pKa = 5.07GEE425 pKa = 4.24DD426 pKa = 3.67HH427 pKa = 6.82TPGDD431 pKa = 3.32IDD433 pKa = 3.35GRR435 pKa = 11.84MAIFNASYY443 pKa = 10.97DD444 pKa = 3.4PGVFYY449 pKa = 9.85TASITGALPNVPITYY464 pKa = 10.05DD465 pKa = 3.51FWVLNIDD472 pKa = 3.77RR473 pKa = 11.84TDD475 pKa = 3.58APCVDD480 pKa = 4.39GGSTCTGPGVDD491 pKa = 4.4GSRR494 pKa = 11.84LRR496 pKa = 11.84PNVLVEE502 pKa = 4.46FRR504 pKa = 11.84DD505 pKa = 3.58LTGNVLASITTGDD518 pKa = 3.65IDD520 pKa = 3.9PTPVGDD526 pKa = 4.42TAGNWMNFRR535 pKa = 11.84ADD537 pKa = 3.26LVLNVSQFEE546 pKa = 4.46VVFINNEE553 pKa = 4.0TGGLGNDD560 pKa = 4.49LALDD564 pKa = 4.65DD565 pKa = 4.68IKK567 pKa = 10.81IEE569 pKa = 4.02QTLCDD574 pKa = 3.73VDD576 pKa = 3.56NDD578 pKa = 4.68GIADD582 pKa = 3.95VFDD585 pKa = 5.64LDD587 pKa = 4.56SDD589 pKa = 4.03DD590 pKa = 5.9DD591 pKa = 5.29GIVDD595 pKa = 5.61LIEE598 pKa = 4.47VGLGALNNGNGVIDD612 pKa = 4.34PVVGWVDD619 pKa = 3.44VNGDD623 pKa = 4.0GLHH626 pKa = 7.0DD627 pKa = 3.87AAAGNVIPDD636 pKa = 3.71TDD638 pKa = 4.01GDD640 pKa = 4.08GTPDD644 pKa = 3.88YY645 pKa = 11.4LDD647 pKa = 5.04LDD649 pKa = 4.21SDD651 pKa = 3.77NDD653 pKa = 4.41SIFDD657 pKa = 3.62VDD659 pKa = 4.19EE660 pKa = 4.84SGATNSSMPGFINEE674 pKa = 4.85DD675 pKa = 3.05GDD677 pKa = 4.03INGDD681 pKa = 3.34GTGNNTEE688 pKa = 4.07TEE690 pKa = 4.07AFRR693 pKa = 11.84EE694 pKa = 4.03KK695 pKa = 10.6DD696 pKa = 3.09TDD698 pKa = 3.66GDD700 pKa = 4.13GTSEE704 pKa = 4.48YY705 pKa = 10.59YY706 pKa = 10.93GDD708 pKa = 5.26GILDD712 pKa = 3.71IYY714 pKa = 9.83EE715 pKa = 4.59HH716 pKa = 7.35SSGLYY721 pKa = 10.7GNANQTNPIDD731 pKa = 4.08SDD733 pKa = 3.76GDD735 pKa = 4.03TIPDD739 pKa = 3.68YY740 pKa = 11.45LDD742 pKa = 3.47PTTNTTHH749 pKa = 7.73DD750 pKa = 3.37ISTTLYY756 pKa = 11.16ASLDD760 pKa = 3.78ANNDD764 pKa = 3.88GIIDD768 pKa = 3.91DD769 pKa = 5.11TIDD772 pKa = 3.34TDD774 pKa = 4.09GDD776 pKa = 4.47GILDD780 pKa = 4.93LYY782 pKa = 10.66DD783 pKa = 3.79TDD785 pKa = 5.71DD786 pKa = 3.96MLFGSPRR793 pKa = 11.84DD794 pKa = 3.63LDD796 pKa = 3.43RR797 pKa = 11.84SLYY800 pKa = 10.18IDD802 pKa = 3.54FDD804 pKa = 3.56GRR806 pKa = 11.84NDD808 pKa = 3.65YY809 pKa = 11.26AQDD812 pKa = 3.37TSLLGGLTDD821 pKa = 3.65ATIMAWINIDD831 pKa = 4.93DD832 pKa = 4.42LFSNQGFIVGQNNFNLKK849 pKa = 9.91INNSRR854 pKa = 11.84NLIATANGTTLSNSTVLNTSQWIHH878 pKa = 4.8VAAVYY883 pKa = 10.75SSGTNTFNLYY893 pKa = 10.89VNGEE897 pKa = 4.28EE898 pKa = 4.82VNTTTAASALNVDD911 pKa = 4.01SSLLTLAKK919 pKa = 10.58NPSTDD924 pKa = 2.93TEE926 pKa = 4.48YY927 pKa = 11.05FNGGIEE933 pKa = 3.99EE934 pKa = 4.16VKK936 pKa = 10.54IFGIALTEE944 pKa = 4.0LQLQKK949 pKa = 10.18MVYY952 pKa = 10.13QKK954 pKa = 10.39IEE956 pKa = 4.38DD957 pKa = 3.55NGQILGEE964 pKa = 4.83IIPIDD969 pKa = 4.13IDD971 pKa = 4.26VAWLDD976 pKa = 3.31LLRR979 pKa = 11.84YY980 pKa = 9.76YY981 pKa = 11.2RR982 pKa = 11.84MDD984 pKa = 4.14AYY986 pKa = 11.04QDD988 pKa = 3.96DD989 pKa = 4.44VVDD992 pKa = 4.71NYY994 pKa = 7.81TTSIIDD1000 pKa = 3.37QYY1002 pKa = 10.53PGTFARR1008 pKa = 11.84CYY1010 pKa = 8.62NVKK1013 pKa = 10.29NIKK1016 pKa = 10.23VEE1018 pKa = 4.04TAPMPFVTQAVPSNYY1033 pKa = 10.01DD1034 pKa = 2.58IGVAVSQNNDD1044 pKa = 2.64VRR1046 pKa = 11.84GDD1048 pKa = 3.74DD1049 pKa = 3.59VKK1051 pKa = 11.36DD1052 pKa = 3.67YY1053 pKa = 10.8DD1054 pKa = 3.64WSITHH1059 pKa = 6.52IRR1061 pKa = 11.84HH1062 pKa = 7.18DD1063 pKa = 3.56INLDD1067 pKa = 3.65DD1068 pKa = 4.04FHH1070 pKa = 9.09SDD1072 pKa = 3.2LGLMIDD1078 pKa = 3.62TGVLVNLNNDD1088 pKa = 3.4TALQNNWYY1096 pKa = 9.98LNLDD1100 pKa = 3.53GVVDD1104 pKa = 5.07LNGEE1108 pKa = 4.2SQLVQTSEE1116 pKa = 4.57SILDD1120 pKa = 3.64IDD1122 pKa = 3.79SEE1124 pKa = 5.15GYY1126 pKa = 9.68IEE1128 pKa = 5.92RR1129 pKa = 11.84DD1130 pKa = 3.3QQGTANSFTYY1140 pKa = 10.21NYY1142 pKa = 8.07WSSPVSVIGAGVNNTPFSISDD1163 pKa = 3.5VLRR1166 pKa = 11.84DD1167 pKa = 3.77GTTSSTPINLDD1178 pKa = 2.81FDD1180 pKa = 4.23TATTLSTADD1189 pKa = 3.94PYY1191 pKa = 11.73YY1192 pKa = 11.13SDD1194 pKa = 4.61GALTNPRR1201 pKa = 11.84KK1202 pKa = 9.51IASYY1206 pKa = 9.31WLWKK1210 pKa = 10.05FVNQTNDD1217 pKa = 3.05YY1218 pKa = 11.16ANWEE1222 pKa = 4.0WFGAYY1227 pKa = 10.43NNINVADD1234 pKa = 4.32GFSMKK1239 pKa = 10.22GVSSTSSVTDD1249 pKa = 3.11SQNYY1253 pKa = 8.07VFVGKK1258 pKa = 9.53PNNAPNVTGEE1268 pKa = 4.2IVHH1271 pKa = 5.36TTFPGGTSIEE1281 pKa = 4.53GYY1283 pKa = 8.84TYY1285 pKa = 11.38NSLTGNPFPSAMDD1298 pKa = 3.8ADD1300 pKa = 4.48QFILDD1305 pKa = 4.87NISSTTGTIYY1315 pKa = 10.31FWEE1318 pKa = 4.16HH1319 pKa = 5.36WGGGNHH1325 pKa = 5.69NWEE1328 pKa = 5.03DD1329 pKa = 3.56YY1330 pKa = 9.9RR1331 pKa = 11.84AGYY1334 pKa = 10.61SMYY1337 pKa = 10.08TMSGGLPGVSHH1348 pKa = 7.88PDD1350 pKa = 3.26GANVGGGTKK1359 pKa = 9.69TPGQYY1364 pKa = 9.96IPVGQGFYY1372 pKa = 10.19IVSSNAGGDD1381 pKa = 3.64VVFKK1385 pKa = 10.66NSQRR1389 pKa = 11.84IFKK1392 pKa = 10.34IEE1394 pKa = 4.46LDD1396 pKa = 3.85DD1397 pKa = 4.68DD1398 pKa = 4.37TNSDD1402 pKa = 2.99HH1403 pKa = 7.99SIFTRR1408 pKa = 11.84AASKK1412 pKa = 11.28NNINEE1417 pKa = 4.0ASTEE1421 pKa = 4.13RR1422 pKa = 11.84NNNTTNEE1429 pKa = 4.03KK1430 pKa = 9.88QRR1432 pKa = 11.84IRR1434 pKa = 11.84LGFEE1438 pKa = 3.92SPDD1441 pKa = 3.41GYY1443 pKa = 10.86HH1444 pKa = 6.08RR1445 pKa = 11.84QVLMAFLEE1453 pKa = 4.76GATDD1457 pKa = 5.43AIDD1460 pKa = 3.72PGYY1463 pKa = 10.76DD1464 pKa = 3.46GEE1466 pKa = 6.08AGDD1469 pKa = 4.62YY1470 pKa = 10.71LPNDD1474 pKa = 3.61GFFIQDD1480 pKa = 3.39DD1481 pKa = 3.96KK1482 pKa = 11.59YY1483 pKa = 10.6YY1484 pKa = 10.92AIIAFGEE1491 pKa = 3.95FDD1493 pKa = 4.0EE1494 pKa = 4.51EE1495 pKa = 4.8RR1496 pKa = 11.84EE1497 pKa = 4.15VPISIFIDD1505 pKa = 3.56EE1506 pKa = 5.06ANDD1509 pKa = 3.67GGNQKK1514 pKa = 10.3FMVDD1518 pKa = 3.87RR1519 pKa = 11.84IEE1521 pKa = 4.63NMPEE1525 pKa = 3.85TISIYY1530 pKa = 10.5IKK1532 pKa = 10.83DD1533 pKa = 3.87NEE1535 pKa = 4.38TGILHH1540 pKa = 7.36DD1541 pKa = 3.42IKK1543 pKa = 11.56NNVFEE1548 pKa = 4.56VALSTGEE1555 pKa = 3.93HH1556 pKa = 5.29KK1557 pKa = 9.66TRR1559 pKa = 11.84FSVVFQDD1566 pKa = 5.16RR1567 pKa = 11.84VLSLNAEE1574 pKa = 4.19LAEE1577 pKa = 4.45SNDD1580 pKa = 3.15LKK1582 pKa = 11.57VFMNNSNSEE1591 pKa = 3.71IQIINNKK1598 pKa = 8.63NLEE1601 pKa = 4.24LNEE1604 pKa = 3.69ISLYY1608 pKa = 10.96NSIGQTINSWNHH1620 pKa = 6.03LGNDD1624 pKa = 3.59MMISLPLNNISSGVYY1639 pKa = 9.33IVTIKK1644 pKa = 6.46TTKK1647 pKa = 10.42GVVSQKK1653 pKa = 10.63IIIKK1657 pKa = 10.26
MM1 pKa = 6.91VFSVGFSQTVIYY13 pKa = 10.95SEE15 pKa = 4.92DD16 pKa = 3.57FEE18 pKa = 6.27SNIGTWTQDD27 pKa = 3.09TADD30 pKa = 3.74TLDD33 pKa = 2.92WSRR36 pKa = 11.84NSGGTGSGGTGPNNSSDD53 pKa = 3.12GSFYY57 pKa = 10.3MYY59 pKa = 10.59TEE61 pKa = 4.29ASSNYY66 pKa = 7.75NTTANFISEE75 pKa = 4.41PFNLSGISSPIFTFHH90 pKa = 5.98YY91 pKa = 10.82HH92 pKa = 5.16MFGSNMGTLNVNISTDD108 pKa = 3.24GGSTFGAPIWTQNGQHH124 pKa = 5.86QGSNSAAWQQVDD136 pKa = 3.81IDD138 pKa = 4.0LSTYY142 pKa = 10.17IGQTINIQFNGQVGSSYY159 pKa = 10.81RR160 pKa = 11.84SDD162 pKa = 3.0IAIDD166 pKa = 4.36EE167 pKa = 4.38ISLTVTSSPDD177 pKa = 3.28PEE179 pKa = 4.39IDD181 pKa = 3.39VTGNSLPIIGDD192 pKa = 3.48GTNAPSINDD201 pKa = 3.26NTDD204 pKa = 3.13FGTVQVGSEE213 pKa = 4.21TSIKK217 pKa = 10.06TYY219 pKa = 10.37TITNNGSLDD228 pKa = 3.8LVLGTSTISGSSDD241 pKa = 3.2FVISTFPGTTITPGNSEE258 pKa = 4.04IIEE261 pKa = 4.02ISFNTLSLGIQTAQLSIVNNDD282 pKa = 3.54PDD284 pKa = 4.02EE285 pKa = 4.43NPYY288 pKa = 10.63LINLSAEE295 pKa = 4.27GTQAFYY301 pKa = 11.09DD302 pKa = 3.71SDD304 pKa = 4.18GDD306 pKa = 4.43GVFDD310 pKa = 6.2DD311 pKa = 5.63VDD313 pKa = 3.9IDD315 pKa = 4.48DD316 pKa = 5.6DD317 pKa = 4.34NDD319 pKa = 4.71GIIDD323 pKa = 3.77SLEE326 pKa = 3.9EE327 pKa = 4.53SNCSGSIITNSVNYY341 pKa = 10.05KK342 pKa = 10.08FLNEE346 pKa = 4.05TFGTGGRR353 pKa = 11.84TTINTSYY360 pKa = 11.18NATTTYY366 pKa = 10.31CYY368 pKa = 10.68EE369 pKa = 5.06DD370 pKa = 3.7GTAGTDD376 pKa = 3.36TTGCPTLSDD385 pKa = 3.31TSLNDD390 pKa = 3.36GEE392 pKa = 4.5YY393 pKa = 9.72TVYY396 pKa = 10.99SRR398 pKa = 11.84ITNNDD403 pKa = 3.07DD404 pKa = 3.16VADD407 pKa = 5.25GINVDD412 pKa = 3.33IASWAEE418 pKa = 3.85DD419 pKa = 3.37FWYY422 pKa = 10.09EE423 pKa = 5.07GEE425 pKa = 4.24DD426 pKa = 3.67HH427 pKa = 6.82TPGDD431 pKa = 3.32IDD433 pKa = 3.35GRR435 pKa = 11.84MAIFNASYY443 pKa = 10.97DD444 pKa = 3.4PGVFYY449 pKa = 9.85TASITGALPNVPITYY464 pKa = 10.05DD465 pKa = 3.51FWVLNIDD472 pKa = 3.77RR473 pKa = 11.84TDD475 pKa = 3.58APCVDD480 pKa = 4.39GGSTCTGPGVDD491 pKa = 4.4GSRR494 pKa = 11.84LRR496 pKa = 11.84PNVLVEE502 pKa = 4.46FRR504 pKa = 11.84DD505 pKa = 3.58LTGNVLASITTGDD518 pKa = 3.65IDD520 pKa = 3.9PTPVGDD526 pKa = 4.42TAGNWMNFRR535 pKa = 11.84ADD537 pKa = 3.26LVLNVSQFEE546 pKa = 4.46VVFINNEE553 pKa = 4.0TGGLGNDD560 pKa = 4.49LALDD564 pKa = 4.65DD565 pKa = 4.68IKK567 pKa = 10.81IEE569 pKa = 4.02QTLCDD574 pKa = 3.73VDD576 pKa = 3.56NDD578 pKa = 4.68GIADD582 pKa = 3.95VFDD585 pKa = 5.64LDD587 pKa = 4.56SDD589 pKa = 4.03DD590 pKa = 5.9DD591 pKa = 5.29GIVDD595 pKa = 5.61LIEE598 pKa = 4.47VGLGALNNGNGVIDD612 pKa = 4.34PVVGWVDD619 pKa = 3.44VNGDD623 pKa = 4.0GLHH626 pKa = 7.0DD627 pKa = 3.87AAAGNVIPDD636 pKa = 3.71TDD638 pKa = 4.01GDD640 pKa = 4.08GTPDD644 pKa = 3.88YY645 pKa = 11.4LDD647 pKa = 5.04LDD649 pKa = 4.21SDD651 pKa = 3.77NDD653 pKa = 4.41SIFDD657 pKa = 3.62VDD659 pKa = 4.19EE660 pKa = 4.84SGATNSSMPGFINEE674 pKa = 4.85DD675 pKa = 3.05GDD677 pKa = 4.03INGDD681 pKa = 3.34GTGNNTEE688 pKa = 4.07TEE690 pKa = 4.07AFRR693 pKa = 11.84EE694 pKa = 4.03KK695 pKa = 10.6DD696 pKa = 3.09TDD698 pKa = 3.66GDD700 pKa = 4.13GTSEE704 pKa = 4.48YY705 pKa = 10.59YY706 pKa = 10.93GDD708 pKa = 5.26GILDD712 pKa = 3.71IYY714 pKa = 9.83EE715 pKa = 4.59HH716 pKa = 7.35SSGLYY721 pKa = 10.7GNANQTNPIDD731 pKa = 4.08SDD733 pKa = 3.76GDD735 pKa = 4.03TIPDD739 pKa = 3.68YY740 pKa = 11.45LDD742 pKa = 3.47PTTNTTHH749 pKa = 7.73DD750 pKa = 3.37ISTTLYY756 pKa = 11.16ASLDD760 pKa = 3.78ANNDD764 pKa = 3.88GIIDD768 pKa = 3.91DD769 pKa = 5.11TIDD772 pKa = 3.34TDD774 pKa = 4.09GDD776 pKa = 4.47GILDD780 pKa = 4.93LYY782 pKa = 10.66DD783 pKa = 3.79TDD785 pKa = 5.71DD786 pKa = 3.96MLFGSPRR793 pKa = 11.84DD794 pKa = 3.63LDD796 pKa = 3.43RR797 pKa = 11.84SLYY800 pKa = 10.18IDD802 pKa = 3.54FDD804 pKa = 3.56GRR806 pKa = 11.84NDD808 pKa = 3.65YY809 pKa = 11.26AQDD812 pKa = 3.37TSLLGGLTDD821 pKa = 3.65ATIMAWINIDD831 pKa = 4.93DD832 pKa = 4.42LFSNQGFIVGQNNFNLKK849 pKa = 9.91INNSRR854 pKa = 11.84NLIATANGTTLSNSTVLNTSQWIHH878 pKa = 4.8VAAVYY883 pKa = 10.75SSGTNTFNLYY893 pKa = 10.89VNGEE897 pKa = 4.28EE898 pKa = 4.82VNTTTAASALNVDD911 pKa = 4.01SSLLTLAKK919 pKa = 10.58NPSTDD924 pKa = 2.93TEE926 pKa = 4.48YY927 pKa = 11.05FNGGIEE933 pKa = 3.99EE934 pKa = 4.16VKK936 pKa = 10.54IFGIALTEE944 pKa = 4.0LQLQKK949 pKa = 10.18MVYY952 pKa = 10.13QKK954 pKa = 10.39IEE956 pKa = 4.38DD957 pKa = 3.55NGQILGEE964 pKa = 4.83IIPIDD969 pKa = 4.13IDD971 pKa = 4.26VAWLDD976 pKa = 3.31LLRR979 pKa = 11.84YY980 pKa = 9.76YY981 pKa = 11.2RR982 pKa = 11.84MDD984 pKa = 4.14AYY986 pKa = 11.04QDD988 pKa = 3.96DD989 pKa = 4.44VVDD992 pKa = 4.71NYY994 pKa = 7.81TTSIIDD1000 pKa = 3.37QYY1002 pKa = 10.53PGTFARR1008 pKa = 11.84CYY1010 pKa = 8.62NVKK1013 pKa = 10.29NIKK1016 pKa = 10.23VEE1018 pKa = 4.04TAPMPFVTQAVPSNYY1033 pKa = 10.01DD1034 pKa = 2.58IGVAVSQNNDD1044 pKa = 2.64VRR1046 pKa = 11.84GDD1048 pKa = 3.74DD1049 pKa = 3.59VKK1051 pKa = 11.36DD1052 pKa = 3.67YY1053 pKa = 10.8DD1054 pKa = 3.64WSITHH1059 pKa = 6.52IRR1061 pKa = 11.84HH1062 pKa = 7.18DD1063 pKa = 3.56INLDD1067 pKa = 3.65DD1068 pKa = 4.04FHH1070 pKa = 9.09SDD1072 pKa = 3.2LGLMIDD1078 pKa = 3.62TGVLVNLNNDD1088 pKa = 3.4TALQNNWYY1096 pKa = 9.98LNLDD1100 pKa = 3.53GVVDD1104 pKa = 5.07LNGEE1108 pKa = 4.2SQLVQTSEE1116 pKa = 4.57SILDD1120 pKa = 3.64IDD1122 pKa = 3.79SEE1124 pKa = 5.15GYY1126 pKa = 9.68IEE1128 pKa = 5.92RR1129 pKa = 11.84DD1130 pKa = 3.3QQGTANSFTYY1140 pKa = 10.21NYY1142 pKa = 8.07WSSPVSVIGAGVNNTPFSISDD1163 pKa = 3.5VLRR1166 pKa = 11.84DD1167 pKa = 3.77GTTSSTPINLDD1178 pKa = 2.81FDD1180 pKa = 4.23TATTLSTADD1189 pKa = 3.94PYY1191 pKa = 11.73YY1192 pKa = 11.13SDD1194 pKa = 4.61GALTNPRR1201 pKa = 11.84KK1202 pKa = 9.51IASYY1206 pKa = 9.31WLWKK1210 pKa = 10.05FVNQTNDD1217 pKa = 3.05YY1218 pKa = 11.16ANWEE1222 pKa = 4.0WFGAYY1227 pKa = 10.43NNINVADD1234 pKa = 4.32GFSMKK1239 pKa = 10.22GVSSTSSVTDD1249 pKa = 3.11SQNYY1253 pKa = 8.07VFVGKK1258 pKa = 9.53PNNAPNVTGEE1268 pKa = 4.2IVHH1271 pKa = 5.36TTFPGGTSIEE1281 pKa = 4.53GYY1283 pKa = 8.84TYY1285 pKa = 11.38NSLTGNPFPSAMDD1298 pKa = 3.8ADD1300 pKa = 4.48QFILDD1305 pKa = 4.87NISSTTGTIYY1315 pKa = 10.31FWEE1318 pKa = 4.16HH1319 pKa = 5.36WGGGNHH1325 pKa = 5.69NWEE1328 pKa = 5.03DD1329 pKa = 3.56YY1330 pKa = 9.9RR1331 pKa = 11.84AGYY1334 pKa = 10.61SMYY1337 pKa = 10.08TMSGGLPGVSHH1348 pKa = 7.88PDD1350 pKa = 3.26GANVGGGTKK1359 pKa = 9.69TPGQYY1364 pKa = 9.96IPVGQGFYY1372 pKa = 10.19IVSSNAGGDD1381 pKa = 3.64VVFKK1385 pKa = 10.66NSQRR1389 pKa = 11.84IFKK1392 pKa = 10.34IEE1394 pKa = 4.46LDD1396 pKa = 3.85DD1397 pKa = 4.68DD1398 pKa = 4.37TNSDD1402 pKa = 2.99HH1403 pKa = 7.99SIFTRR1408 pKa = 11.84AASKK1412 pKa = 11.28NNINEE1417 pKa = 4.0ASTEE1421 pKa = 4.13RR1422 pKa = 11.84NNNTTNEE1429 pKa = 4.03KK1430 pKa = 9.88QRR1432 pKa = 11.84IRR1434 pKa = 11.84LGFEE1438 pKa = 3.92SPDD1441 pKa = 3.41GYY1443 pKa = 10.86HH1444 pKa = 6.08RR1445 pKa = 11.84QVLMAFLEE1453 pKa = 4.76GATDD1457 pKa = 5.43AIDD1460 pKa = 3.72PGYY1463 pKa = 10.76DD1464 pKa = 3.46GEE1466 pKa = 6.08AGDD1469 pKa = 4.62YY1470 pKa = 10.71LPNDD1474 pKa = 3.61GFFIQDD1480 pKa = 3.39DD1481 pKa = 3.96KK1482 pKa = 11.59YY1483 pKa = 10.6YY1484 pKa = 10.92AIIAFGEE1491 pKa = 3.95FDD1493 pKa = 4.0EE1494 pKa = 4.51EE1495 pKa = 4.8RR1496 pKa = 11.84EE1497 pKa = 4.15VPISIFIDD1505 pKa = 3.56EE1506 pKa = 5.06ANDD1509 pKa = 3.67GGNQKK1514 pKa = 10.3FMVDD1518 pKa = 3.87RR1519 pKa = 11.84IEE1521 pKa = 4.63NMPEE1525 pKa = 3.85TISIYY1530 pKa = 10.5IKK1532 pKa = 10.83DD1533 pKa = 3.87NEE1535 pKa = 4.38TGILHH1540 pKa = 7.36DD1541 pKa = 3.42IKK1543 pKa = 11.56NNVFEE1548 pKa = 4.56VALSTGEE1555 pKa = 3.93HH1556 pKa = 5.29KK1557 pKa = 9.66TRR1559 pKa = 11.84FSVVFQDD1566 pKa = 5.16RR1567 pKa = 11.84VLSLNAEE1574 pKa = 4.19LAEE1577 pKa = 4.45SNDD1580 pKa = 3.15LKK1582 pKa = 11.57VFMNNSNSEE1591 pKa = 3.71IQIINNKK1598 pKa = 8.63NLEE1601 pKa = 4.24LNEE1604 pKa = 3.69ISLYY1608 pKa = 10.96NSIGQTINSWNHH1620 pKa = 6.03LGNDD1624 pKa = 3.59MMISLPLNNISSGVYY1639 pKa = 9.33IVTIKK1644 pKa = 6.46TTKK1647 pKa = 10.42GVVSQKK1653 pKa = 10.63IIIKK1657 pKa = 10.26
Molecular weight: 179.9 kDa
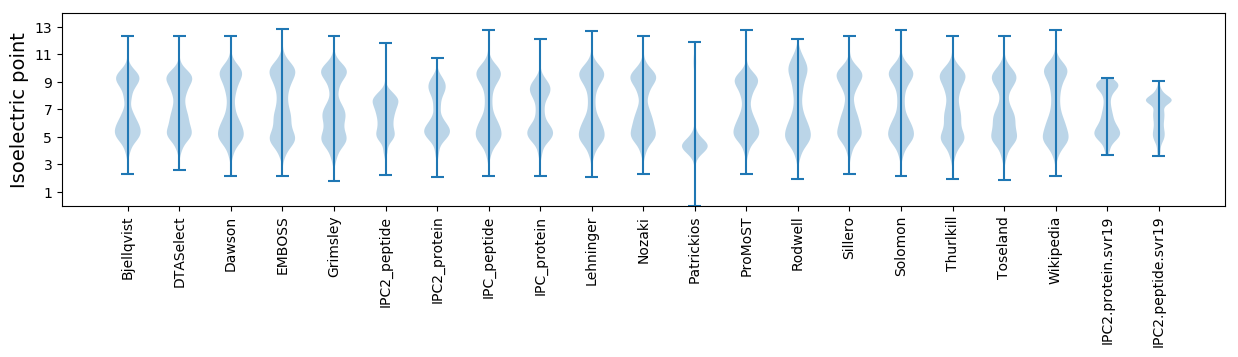
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D8PBP7|A0A1D8PBP7_9FLAO PDZ domain-containing protein OS=Urechidicola croceus OX=1850246 GN=LPB138_04700 PE=4 SV=1
MM1 pKa = 7.63KK2 pKa = 10.41KK3 pKa = 10.58GILFIAAMLLLVSVSEE19 pKa = 4.27ANNNGRR25 pKa = 11.84INSRR29 pKa = 11.84MGYY32 pKa = 7.83TNVYY36 pKa = 9.37DD37 pKa = 3.85AQPIQFKK44 pKa = 10.64EE45 pKa = 3.99KK46 pKa = 10.38GIQFYY51 pKa = 11.33VFLDD55 pKa = 3.93GQFDD59 pKa = 4.51FSTRR63 pKa = 11.84PNRR66 pKa = 11.84NFNNQYY72 pKa = 10.98KK73 pKa = 10.15NLNSRR78 pKa = 11.84RR79 pKa = 11.84NLRR82 pKa = 11.84IINNRR87 pKa = 11.84GLRR90 pKa = 11.84IEE92 pKa = 3.89RR93 pKa = 11.84DD94 pKa = 3.13YY95 pKa = 11.07RR96 pKa = 11.84GRR98 pKa = 11.84IRR100 pKa = 11.84RR101 pKa = 11.84VGNVPINYY109 pKa = 8.73NRR111 pKa = 11.84LGKK114 pKa = 7.85VTRR117 pKa = 11.84IGSVFIGYY125 pKa = 8.87DD126 pKa = 3.47FRR128 pKa = 11.84QVSQVGGLNVLYY140 pKa = 9.99NHH142 pKa = 7.27WGDD145 pKa = 3.14ARR147 pKa = 11.84YY148 pKa = 9.69IGQVKK153 pKa = 9.72YY154 pKa = 10.94NNHH157 pKa = 6.0RR158 pKa = 11.84FNGNFSVYY166 pKa = 9.03NTWNTWEE173 pKa = 4.04YY174 pKa = 11.52DD175 pKa = 3.54FNNDD179 pKa = 3.01FFLRR183 pKa = 11.84SNFRR187 pKa = 11.84NDD189 pKa = 3.0FEE191 pKa = 5.62SFHH194 pKa = 7.07EE195 pKa = 4.65DD196 pKa = 3.94DD197 pKa = 3.58NYY199 pKa = 11.38LYY201 pKa = 10.77YY202 pKa = 10.39RR203 pKa = 11.84SKK205 pKa = 10.98GVKK208 pKa = 9.27KK209 pKa = 10.32GKK211 pKa = 9.39KK212 pKa = 9.34GNVIKK217 pKa = 10.67RR218 pKa = 11.84KK219 pKa = 9.54KK220 pKa = 9.87NKK222 pKa = 9.89QKK224 pKa = 10.45VFPNKK229 pKa = 7.86NTRR232 pKa = 11.84RR233 pKa = 3.75
MM1 pKa = 7.63KK2 pKa = 10.41KK3 pKa = 10.58GILFIAAMLLLVSVSEE19 pKa = 4.27ANNNGRR25 pKa = 11.84INSRR29 pKa = 11.84MGYY32 pKa = 7.83TNVYY36 pKa = 9.37DD37 pKa = 3.85AQPIQFKK44 pKa = 10.64EE45 pKa = 3.99KK46 pKa = 10.38GIQFYY51 pKa = 11.33VFLDD55 pKa = 3.93GQFDD59 pKa = 4.51FSTRR63 pKa = 11.84PNRR66 pKa = 11.84NFNNQYY72 pKa = 10.98KK73 pKa = 10.15NLNSRR78 pKa = 11.84RR79 pKa = 11.84NLRR82 pKa = 11.84IINNRR87 pKa = 11.84GLRR90 pKa = 11.84IEE92 pKa = 3.89RR93 pKa = 11.84DD94 pKa = 3.13YY95 pKa = 11.07RR96 pKa = 11.84GRR98 pKa = 11.84IRR100 pKa = 11.84RR101 pKa = 11.84VGNVPINYY109 pKa = 8.73NRR111 pKa = 11.84LGKK114 pKa = 7.85VTRR117 pKa = 11.84IGSVFIGYY125 pKa = 8.87DD126 pKa = 3.47FRR128 pKa = 11.84QVSQVGGLNVLYY140 pKa = 9.99NHH142 pKa = 7.27WGDD145 pKa = 3.14ARR147 pKa = 11.84YY148 pKa = 9.69IGQVKK153 pKa = 9.72YY154 pKa = 10.94NNHH157 pKa = 6.0RR158 pKa = 11.84FNGNFSVYY166 pKa = 9.03NTWNTWEE173 pKa = 4.04YY174 pKa = 11.52DD175 pKa = 3.54FNNDD179 pKa = 3.01FFLRR183 pKa = 11.84SNFRR187 pKa = 11.84NDD189 pKa = 3.0FEE191 pKa = 5.62SFHH194 pKa = 7.07EE195 pKa = 4.65DD196 pKa = 3.94DD197 pKa = 3.58NYY199 pKa = 11.38LYY201 pKa = 10.77YY202 pKa = 10.39RR203 pKa = 11.84SKK205 pKa = 10.98GVKK208 pKa = 9.27KK209 pKa = 10.32GKK211 pKa = 9.39KK212 pKa = 9.34GNVIKK217 pKa = 10.67RR218 pKa = 11.84KK219 pKa = 9.54KK220 pKa = 9.87NKK222 pKa = 9.89QKK224 pKa = 10.45VFPNKK229 pKa = 7.86NTRR232 pKa = 11.84RR233 pKa = 3.75
Molecular weight: 27.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1027031 |
49 |
6637 |
348.6 |
39.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.46 ± 0.047 | 0.716 ± 0.015 |
5.856 ± 0.091 | 6.735 ± 0.044 |
5.265 ± 0.033 | 6.241 ± 0.047 |
1.676 ± 0.022 | 8.741 ± 0.042 |
8.196 ± 0.088 | 9.059 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.071 ± 0.027 | 6.798 ± 0.05 |
3.209 ± 0.025 | 3.154 ± 0.028 |
3.113 ± 0.036 | 6.721 ± 0.041 |
5.856 ± 0.09 | 6.045 ± 0.036 |
1.011 ± 0.016 | 4.077 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
