
Rhodovulum bhavnagarense
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Rhodovulum
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
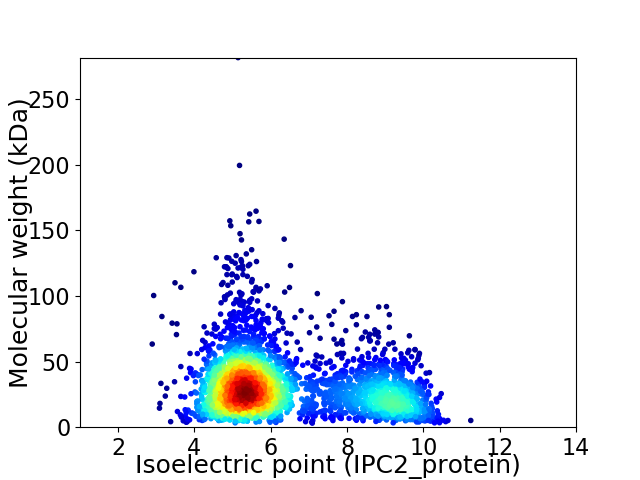
Virtual 2D-PAGE plot for 3085 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R2RH35|A0A4R2RH35_9RHOB Site-specific DNA-methyltransferase (adenine-specific) OS=Rhodovulum bhavnagarense OX=992286 GN=EV663_12416 PE=3 SV=1
MM1 pKa = 7.71TYY3 pKa = 10.7DD4 pKa = 4.9CDD6 pKa = 5.05DD7 pKa = 3.65STPTEE12 pKa = 4.2TANATADD19 pKa = 3.48TSSFAGNVIGLWEE32 pKa = 4.34FDD34 pKa = 4.52DD35 pKa = 4.57GCSYY39 pKa = 10.56YY40 pKa = 11.12ARR42 pKa = 11.84DD43 pKa = 3.44TGLDD47 pKa = 3.43DD48 pKa = 5.49GVAQNGTYY56 pKa = 10.63EE57 pKa = 4.22NGANDD62 pKa = 3.52YY63 pKa = 11.07GGWLTLDD70 pKa = 4.44GYY72 pKa = 11.07NDD74 pKa = 3.78RR75 pKa = 11.84VDD77 pKa = 3.94VDD79 pKa = 4.36GKK81 pKa = 9.5DD82 pKa = 3.36TPFDD86 pKa = 4.0LSAGTIQTRR95 pKa = 11.84FIADD99 pKa = 3.85GSYY102 pKa = 10.45SKK104 pKa = 11.14CGGTQLEE111 pKa = 4.86GGTLVNRR118 pKa = 11.84GEE120 pKa = 4.04YY121 pKa = 9.96ADD123 pKa = 5.36RR124 pKa = 11.84YY125 pKa = 9.8DD126 pKa = 3.28EE127 pKa = 5.43GYY129 pKa = 10.76FGLSTTASGAVMVEE143 pKa = 4.51HH144 pKa = 6.42YY145 pKa = 10.68AAGASVTLSTQDD157 pKa = 2.56GFYY160 pKa = 11.33AMGDD164 pKa = 3.85EE165 pKa = 4.44VTVTYY170 pKa = 10.55SWDD173 pKa = 3.45ADD175 pKa = 3.46GGATLIVLNATTGQTAMLSDD195 pKa = 3.87DD196 pKa = 3.91TVTGLTLDD204 pKa = 3.76IGDD207 pKa = 4.41NDD209 pKa = 4.54DD210 pKa = 3.73EE211 pKa = 4.84NFTFGARR218 pKa = 11.84EE219 pKa = 3.93VDD221 pKa = 3.26DD222 pKa = 5.13GKK224 pKa = 11.4YY225 pKa = 10.51DD226 pKa = 3.4DD227 pKa = 5.51HH228 pKa = 8.52FDD230 pKa = 3.43GKK232 pKa = 10.17IAFVAVYY239 pKa = 8.41DD240 pKa = 3.91QPVAAGDD247 pKa = 3.9GFVEE251 pKa = 5.0GTAGDD256 pKa = 4.23DD257 pKa = 5.01LIDD260 pKa = 3.66AAYY263 pKa = 9.33TGDD266 pKa = 4.17PEE268 pKa = 5.12EE269 pKa = 5.44DD270 pKa = 3.6MIDD273 pKa = 3.59AGDD276 pKa = 4.73AILPGEE282 pKa = 4.84APDD285 pKa = 5.22DD286 pKa = 4.98DD287 pKa = 4.19IVLAGDD293 pKa = 3.83GNDD296 pKa = 3.19TVLAGEE302 pKa = 4.91GDD304 pKa = 3.94DD305 pKa = 3.87EE306 pKa = 5.2VYY308 pKa = 10.88AGTGIDD314 pKa = 3.66EE315 pKa = 4.62VEE317 pKa = 4.74GGTGDD322 pKa = 3.73DD323 pKa = 4.81LIYY326 pKa = 10.71GGSGADD332 pKa = 3.52ILDD335 pKa = 4.05GQAGNDD341 pKa = 3.88TIFGDD346 pKa = 4.04GGNAPTIATITFEE359 pKa = 4.64SEE361 pKa = 3.52DD362 pKa = 3.53AGYY365 pKa = 10.51LNTMGVYY372 pKa = 10.08TIDD375 pKa = 4.78PDD377 pKa = 3.45TGAITNVEE385 pKa = 3.81IAFEE389 pKa = 4.14NTSAIGSGGNLTQGDD404 pKa = 4.81SYY406 pKa = 11.41TYY408 pKa = 9.23VTGPGAQVGVFLIADD423 pKa = 4.83GYY425 pKa = 11.27DD426 pKa = 3.53LNDD429 pKa = 3.4YY430 pKa = 10.37AALGEE435 pKa = 4.34GSFLFLDD442 pKa = 4.15ASGNPATLDD451 pKa = 3.49TPNPSLVFVAEE462 pKa = 5.18DD463 pKa = 3.68GTQTALSGEE472 pKa = 4.27IFHH475 pKa = 6.51TAAHH479 pKa = 5.77GTNLGLNPDD488 pKa = 4.32GLLHH492 pKa = 5.81TAGYY496 pKa = 9.98SEE498 pKa = 4.85NPDD501 pKa = 3.06GSVTLGFEE509 pKa = 5.15DD510 pKa = 4.2INGLGDD516 pKa = 3.53QDD518 pKa = 3.96YY519 pKa = 10.49DD520 pKa = 4.11DD521 pKa = 4.87PVFTVAIEE529 pKa = 4.22GAGGSLNNAHH539 pKa = 7.14FDD541 pKa = 3.93LEE543 pKa = 4.37NPADD547 pKa = 3.88ILTEE551 pKa = 4.38GDD553 pKa = 3.6DD554 pKa = 3.96SLIGGDD560 pKa = 3.9GADD563 pKa = 3.51VIYY566 pKa = 10.75GGGGNDD572 pKa = 3.86TIHH575 pKa = 6.98AGSGLTSQSPDD586 pKa = 3.01LGYY589 pKa = 10.02PGLFDD594 pKa = 6.27ADD596 pKa = 3.97TDD598 pKa = 4.14PEE600 pKa = 4.39DD601 pKa = 4.72DD602 pKa = 3.73RR603 pKa = 11.84DD604 pKa = 4.08YY605 pKa = 12.0VDD607 pKa = 5.42GGDD610 pKa = 3.93GNDD613 pKa = 5.31LITTGDD619 pKa = 4.0DD620 pKa = 3.21ADD622 pKa = 4.25TIYY625 pKa = 11.19GGTGNDD631 pKa = 3.77TIDD634 pKa = 3.63AGVDD638 pKa = 2.85ADD640 pKa = 4.96EE641 pKa = 4.39IHH643 pKa = 6.92GGAGDD648 pKa = 3.81DD649 pKa = 4.4SIIGGEE655 pKa = 4.24GSDD658 pKa = 4.21TIAGGAGDD666 pKa = 3.73DD667 pKa = 4.17TIYY670 pKa = 11.27GGLDD674 pKa = 3.16PAYY677 pKa = 9.11PDD679 pKa = 4.14SLNIPDD685 pKa = 4.22NTDD688 pKa = 3.39LVPDD692 pKa = 4.07NGRR695 pKa = 11.84DD696 pKa = 3.73LIDD699 pKa = 4.48GGDD702 pKa = 4.07GNDD705 pKa = 4.76LIYY708 pKa = 10.85GQDD711 pKa = 4.11DD712 pKa = 4.32DD713 pKa = 3.99DD714 pKa = 4.68TIYY717 pKa = 11.26GGAGNDD723 pKa = 4.08TIDD726 pKa = 4.47AGIDD730 pKa = 3.32DD731 pKa = 5.46DD732 pKa = 4.86YY733 pKa = 11.98VDD735 pKa = 4.2GGEE738 pKa = 4.28GADD741 pKa = 3.87VIYY744 pKa = 10.85SGAGSDD750 pKa = 3.85TIEE753 pKa = 5.12GGDD756 pKa = 4.12GDD758 pKa = 4.18DD759 pKa = 4.54TIHH762 pKa = 7.2GGGDD766 pKa = 3.44NDD768 pKa = 4.02VLSGGADD775 pKa = 3.18RR776 pKa = 11.84DD777 pKa = 3.19IFYY780 pKa = 10.0IDD782 pKa = 3.56SVGMPGGSTNTTVHH796 pKa = 6.39GGNTGVDD803 pKa = 3.24WDD805 pKa = 4.37TLNLSALISGGWTITHH821 pKa = 6.9LVKK824 pKa = 11.09NPDD827 pKa = 3.64GDD829 pKa = 4.25PDD831 pKa = 3.93GLGWDD836 pKa = 4.14GQITLQDD843 pKa = 3.17GGGQYY848 pKa = 11.66ANINFTNIEE857 pKa = 4.13EE858 pKa = 4.73IIPCFTPGTLIATPKK873 pKa = 10.31GEE875 pKa = 4.09RR876 pKa = 11.84PVEE879 pKa = 3.86EE880 pKa = 4.44LRR882 pKa = 11.84EE883 pKa = 3.95GDD885 pKa = 4.32KK886 pKa = 11.27VITRR890 pKa = 11.84DD891 pKa = 3.49NGIQEE896 pKa = 4.13IRR898 pKa = 11.84WIGTTNVSSEE908 pKa = 3.84RR909 pKa = 11.84LAHH912 pKa = 6.86DD913 pKa = 3.43KK914 pKa = 11.1HH915 pKa = 7.17LMPVLIRR922 pKa = 11.84AGALGHH928 pKa = 6.3GLPEE932 pKa = 5.05RR933 pKa = 11.84DD934 pKa = 3.33MLVSPQHH941 pKa = 6.0RR942 pKa = 11.84VLVANDD948 pKa = 3.21RR949 pKa = 11.84TALYY953 pKa = 10.34FEE955 pKa = 4.27EE956 pKa = 5.98RR957 pKa = 11.84EE958 pKa = 4.11VLVAAKK964 pKa = 10.08HH965 pKa = 5.15LVNNRR970 pKa = 11.84SIQRR974 pKa = 11.84VAPRR978 pKa = 11.84ALSYY982 pKa = 10.79LHH984 pKa = 6.82FMFDD988 pKa = 3.02QHH990 pKa = 6.54EE991 pKa = 4.6VVLSDD996 pKa = 3.99GAWTEE1001 pKa = 4.14SFQPGDD1007 pKa = 3.58YY1008 pKa = 10.27TLGGMGNAQRR1018 pKa = 11.84GEE1020 pKa = 3.95IFEE1023 pKa = 5.23LFPEE1027 pKa = 4.63LQTEE1031 pKa = 4.13EE1032 pKa = 4.27GLRR1035 pKa = 11.84DD1036 pKa = 3.39YY1037 pKa = 11.13VAARR1041 pKa = 11.84RR1042 pKa = 11.84TLRR1045 pKa = 11.84RR1046 pKa = 11.84HH1047 pKa = 5.0EE1048 pKa = 4.08AVLLGSEE1055 pKa = 4.56FF1056 pKa = 3.56
MM1 pKa = 7.71TYY3 pKa = 10.7DD4 pKa = 4.9CDD6 pKa = 5.05DD7 pKa = 3.65STPTEE12 pKa = 4.2TANATADD19 pKa = 3.48TSSFAGNVIGLWEE32 pKa = 4.34FDD34 pKa = 4.52DD35 pKa = 4.57GCSYY39 pKa = 10.56YY40 pKa = 11.12ARR42 pKa = 11.84DD43 pKa = 3.44TGLDD47 pKa = 3.43DD48 pKa = 5.49GVAQNGTYY56 pKa = 10.63EE57 pKa = 4.22NGANDD62 pKa = 3.52YY63 pKa = 11.07GGWLTLDD70 pKa = 4.44GYY72 pKa = 11.07NDD74 pKa = 3.78RR75 pKa = 11.84VDD77 pKa = 3.94VDD79 pKa = 4.36GKK81 pKa = 9.5DD82 pKa = 3.36TPFDD86 pKa = 4.0LSAGTIQTRR95 pKa = 11.84FIADD99 pKa = 3.85GSYY102 pKa = 10.45SKK104 pKa = 11.14CGGTQLEE111 pKa = 4.86GGTLVNRR118 pKa = 11.84GEE120 pKa = 4.04YY121 pKa = 9.96ADD123 pKa = 5.36RR124 pKa = 11.84YY125 pKa = 9.8DD126 pKa = 3.28EE127 pKa = 5.43GYY129 pKa = 10.76FGLSTTASGAVMVEE143 pKa = 4.51HH144 pKa = 6.42YY145 pKa = 10.68AAGASVTLSTQDD157 pKa = 2.56GFYY160 pKa = 11.33AMGDD164 pKa = 3.85EE165 pKa = 4.44VTVTYY170 pKa = 10.55SWDD173 pKa = 3.45ADD175 pKa = 3.46GGATLIVLNATTGQTAMLSDD195 pKa = 3.87DD196 pKa = 3.91TVTGLTLDD204 pKa = 3.76IGDD207 pKa = 4.41NDD209 pKa = 4.54DD210 pKa = 3.73EE211 pKa = 4.84NFTFGARR218 pKa = 11.84EE219 pKa = 3.93VDD221 pKa = 3.26DD222 pKa = 5.13GKK224 pKa = 11.4YY225 pKa = 10.51DD226 pKa = 3.4DD227 pKa = 5.51HH228 pKa = 8.52FDD230 pKa = 3.43GKK232 pKa = 10.17IAFVAVYY239 pKa = 8.41DD240 pKa = 3.91QPVAAGDD247 pKa = 3.9GFVEE251 pKa = 5.0GTAGDD256 pKa = 4.23DD257 pKa = 5.01LIDD260 pKa = 3.66AAYY263 pKa = 9.33TGDD266 pKa = 4.17PEE268 pKa = 5.12EE269 pKa = 5.44DD270 pKa = 3.6MIDD273 pKa = 3.59AGDD276 pKa = 4.73AILPGEE282 pKa = 4.84APDD285 pKa = 5.22DD286 pKa = 4.98DD287 pKa = 4.19IVLAGDD293 pKa = 3.83GNDD296 pKa = 3.19TVLAGEE302 pKa = 4.91GDD304 pKa = 3.94DD305 pKa = 3.87EE306 pKa = 5.2VYY308 pKa = 10.88AGTGIDD314 pKa = 3.66EE315 pKa = 4.62VEE317 pKa = 4.74GGTGDD322 pKa = 3.73DD323 pKa = 4.81LIYY326 pKa = 10.71GGSGADD332 pKa = 3.52ILDD335 pKa = 4.05GQAGNDD341 pKa = 3.88TIFGDD346 pKa = 4.04GGNAPTIATITFEE359 pKa = 4.64SEE361 pKa = 3.52DD362 pKa = 3.53AGYY365 pKa = 10.51LNTMGVYY372 pKa = 10.08TIDD375 pKa = 4.78PDD377 pKa = 3.45TGAITNVEE385 pKa = 3.81IAFEE389 pKa = 4.14NTSAIGSGGNLTQGDD404 pKa = 4.81SYY406 pKa = 11.41TYY408 pKa = 9.23VTGPGAQVGVFLIADD423 pKa = 4.83GYY425 pKa = 11.27DD426 pKa = 3.53LNDD429 pKa = 3.4YY430 pKa = 10.37AALGEE435 pKa = 4.34GSFLFLDD442 pKa = 4.15ASGNPATLDD451 pKa = 3.49TPNPSLVFVAEE462 pKa = 5.18DD463 pKa = 3.68GTQTALSGEE472 pKa = 4.27IFHH475 pKa = 6.51TAAHH479 pKa = 5.77GTNLGLNPDD488 pKa = 4.32GLLHH492 pKa = 5.81TAGYY496 pKa = 9.98SEE498 pKa = 4.85NPDD501 pKa = 3.06GSVTLGFEE509 pKa = 5.15DD510 pKa = 4.2INGLGDD516 pKa = 3.53QDD518 pKa = 3.96YY519 pKa = 10.49DD520 pKa = 4.11DD521 pKa = 4.87PVFTVAIEE529 pKa = 4.22GAGGSLNNAHH539 pKa = 7.14FDD541 pKa = 3.93LEE543 pKa = 4.37NPADD547 pKa = 3.88ILTEE551 pKa = 4.38GDD553 pKa = 3.6DD554 pKa = 3.96SLIGGDD560 pKa = 3.9GADD563 pKa = 3.51VIYY566 pKa = 10.75GGGGNDD572 pKa = 3.86TIHH575 pKa = 6.98AGSGLTSQSPDD586 pKa = 3.01LGYY589 pKa = 10.02PGLFDD594 pKa = 6.27ADD596 pKa = 3.97TDD598 pKa = 4.14PEE600 pKa = 4.39DD601 pKa = 4.72DD602 pKa = 3.73RR603 pKa = 11.84DD604 pKa = 4.08YY605 pKa = 12.0VDD607 pKa = 5.42GGDD610 pKa = 3.93GNDD613 pKa = 5.31LITTGDD619 pKa = 4.0DD620 pKa = 3.21ADD622 pKa = 4.25TIYY625 pKa = 11.19GGTGNDD631 pKa = 3.77TIDD634 pKa = 3.63AGVDD638 pKa = 2.85ADD640 pKa = 4.96EE641 pKa = 4.39IHH643 pKa = 6.92GGAGDD648 pKa = 3.81DD649 pKa = 4.4SIIGGEE655 pKa = 4.24GSDD658 pKa = 4.21TIAGGAGDD666 pKa = 3.73DD667 pKa = 4.17TIYY670 pKa = 11.27GGLDD674 pKa = 3.16PAYY677 pKa = 9.11PDD679 pKa = 4.14SLNIPDD685 pKa = 4.22NTDD688 pKa = 3.39LVPDD692 pKa = 4.07NGRR695 pKa = 11.84DD696 pKa = 3.73LIDD699 pKa = 4.48GGDD702 pKa = 4.07GNDD705 pKa = 4.76LIYY708 pKa = 10.85GQDD711 pKa = 4.11DD712 pKa = 4.32DD713 pKa = 3.99DD714 pKa = 4.68TIYY717 pKa = 11.26GGAGNDD723 pKa = 4.08TIDD726 pKa = 4.47AGIDD730 pKa = 3.32DD731 pKa = 5.46DD732 pKa = 4.86YY733 pKa = 11.98VDD735 pKa = 4.2GGEE738 pKa = 4.28GADD741 pKa = 3.87VIYY744 pKa = 10.85SGAGSDD750 pKa = 3.85TIEE753 pKa = 5.12GGDD756 pKa = 4.12GDD758 pKa = 4.18DD759 pKa = 4.54TIHH762 pKa = 7.2GGGDD766 pKa = 3.44NDD768 pKa = 4.02VLSGGADD775 pKa = 3.18RR776 pKa = 11.84DD777 pKa = 3.19IFYY780 pKa = 10.0IDD782 pKa = 3.56SVGMPGGSTNTTVHH796 pKa = 6.39GGNTGVDD803 pKa = 3.24WDD805 pKa = 4.37TLNLSALISGGWTITHH821 pKa = 6.9LVKK824 pKa = 11.09NPDD827 pKa = 3.64GDD829 pKa = 4.25PDD831 pKa = 3.93GLGWDD836 pKa = 4.14GQITLQDD843 pKa = 3.17GGGQYY848 pKa = 11.66ANINFTNIEE857 pKa = 4.13EE858 pKa = 4.73IIPCFTPGTLIATPKK873 pKa = 10.31GEE875 pKa = 4.09RR876 pKa = 11.84PVEE879 pKa = 3.86EE880 pKa = 4.44LRR882 pKa = 11.84EE883 pKa = 3.95GDD885 pKa = 4.32KK886 pKa = 11.27VITRR890 pKa = 11.84DD891 pKa = 3.49NGIQEE896 pKa = 4.13IRR898 pKa = 11.84WIGTTNVSSEE908 pKa = 3.84RR909 pKa = 11.84LAHH912 pKa = 6.86DD913 pKa = 3.43KK914 pKa = 11.1HH915 pKa = 7.17LMPVLIRR922 pKa = 11.84AGALGHH928 pKa = 6.3GLPEE932 pKa = 5.05RR933 pKa = 11.84DD934 pKa = 3.33MLVSPQHH941 pKa = 6.0RR942 pKa = 11.84VLVANDD948 pKa = 3.21RR949 pKa = 11.84TALYY953 pKa = 10.34FEE955 pKa = 4.27EE956 pKa = 5.98RR957 pKa = 11.84EE958 pKa = 4.11VLVAAKK964 pKa = 10.08HH965 pKa = 5.15LVNNRR970 pKa = 11.84SIQRR974 pKa = 11.84VAPRR978 pKa = 11.84ALSYY982 pKa = 10.79LHH984 pKa = 6.82FMFDD988 pKa = 3.02QHH990 pKa = 6.54EE991 pKa = 4.6VVLSDD996 pKa = 3.99GAWTEE1001 pKa = 4.14SFQPGDD1007 pKa = 3.58YY1008 pKa = 10.27TLGGMGNAQRR1018 pKa = 11.84GEE1020 pKa = 3.95IFEE1023 pKa = 5.23LFPEE1027 pKa = 4.63LQTEE1031 pKa = 4.13EE1032 pKa = 4.27GLRR1035 pKa = 11.84DD1036 pKa = 3.39YY1037 pKa = 11.13VAARR1041 pKa = 11.84RR1042 pKa = 11.84TLRR1045 pKa = 11.84RR1046 pKa = 11.84HH1047 pKa = 5.0EE1048 pKa = 4.08AVLLGSEE1055 pKa = 4.56FF1056 pKa = 3.56
Molecular weight: 110.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R2RCF0|A0A4R2RCF0_9RHOB DNA polymerase IV OS=Rhodovulum bhavnagarense OX=992286 GN=dinB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84AKK41 pKa = 10.7LSAA44 pKa = 4.0
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84AKK41 pKa = 10.7LSAA44 pKa = 4.0
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
951992 |
28 |
2677 |
308.6 |
33.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.078 ± 0.07 | 0.872 ± 0.014 |
6.035 ± 0.043 | 5.968 ± 0.046 |
3.541 ± 0.028 | 8.866 ± 0.05 |
2.053 ± 0.021 | 4.928 ± 0.03 |
2.792 ± 0.035 | 10.307 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.718 ± 0.022 | 2.3 ± 0.021 |
5.349 ± 0.033 | 3.008 ± 0.023 |
7.615 ± 0.046 | 4.622 ± 0.03 |
5.219 ± 0.031 | 7.293 ± 0.038 |
1.343 ± 0.018 | 2.094 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
