
Dethiosulfatibacter aminovorans DSM 17477
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Tissierellia; Tissierellia incertae sedis; Dethiosulfatibacter; Dethiosulfatibacter aminovorans
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
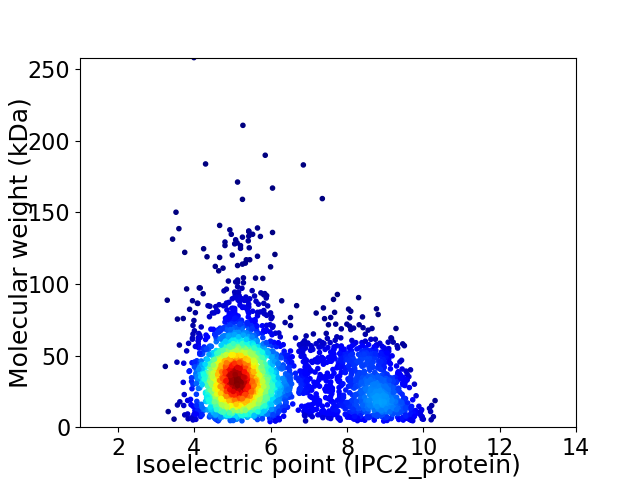
Virtual 2D-PAGE plot for 3628 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6ISX7|A0A1M6ISX7_9FIRM Uncharacterized protein OS=Dethiosulfatibacter aminovorans DSM 17477 OX=1121476 GN=SAMN02745751_02422 PE=4 SV=1
MM1 pKa = 7.19KK2 pKa = 9.94RR3 pKa = 11.84KK4 pKa = 9.23SRR6 pKa = 11.84AVIYY10 pKa = 10.35VILAVILLLSTFVRR24 pKa = 11.84AYY26 pKa = 10.72GDD28 pKa = 3.54YY29 pKa = 9.97GTGNLSSDD37 pKa = 3.58GQQEE41 pKa = 4.38KK42 pKa = 10.6IGITEE47 pKa = 4.01YY48 pKa = 11.3LNDD51 pKa = 3.64QIIVKK56 pKa = 10.17FKK58 pKa = 11.23DD59 pKa = 4.2DD60 pKa = 4.39ISDD63 pKa = 3.58TDD65 pKa = 3.21RR66 pKa = 11.84KK67 pKa = 10.35LIKK70 pKa = 10.01KK71 pKa = 9.4YY72 pKa = 11.02YY73 pKa = 10.28KK74 pKa = 10.7LIDD77 pKa = 3.35KK78 pKa = 10.88AKK80 pKa = 8.51FKK82 pKa = 11.22SGGEE86 pKa = 3.99VLKK89 pKa = 11.2LKK91 pKa = 10.98GNLTVEE97 pKa = 4.66EE98 pKa = 4.22LVEE101 pKa = 3.96MLGEE105 pKa = 4.28LDD107 pKa = 3.65SVEE110 pKa = 4.24YY111 pKa = 10.96AGFDD115 pKa = 3.73YY116 pKa = 10.87IMRR119 pKa = 11.84ATDD122 pKa = 4.03FPDD125 pKa = 4.5DD126 pKa = 3.82PWDD129 pKa = 4.16GDD131 pKa = 3.79SARR134 pKa = 11.84YY135 pKa = 8.16LWGMEE140 pKa = 4.32KK141 pKa = 9.27IDD143 pKa = 3.97ANGDD147 pKa = 3.54TEE149 pKa = 6.03YY150 pKa = 10.78PGAWNLATGNPEE162 pKa = 3.99VIVAVIDD169 pKa = 3.46TGVDD173 pKa = 3.06MGHH176 pKa = 6.49VDD178 pKa = 3.68LAEE181 pKa = 4.56NIWINEE187 pKa = 3.86VEE189 pKa = 4.3LNGDD193 pKa = 3.81AGVDD197 pKa = 3.67DD198 pKa = 5.66DD199 pKa = 5.1GNGYY203 pKa = 9.68IDD205 pKa = 6.01DD206 pKa = 3.63VHH208 pKa = 7.62GWDD211 pKa = 3.81FHH213 pKa = 8.45NGDD216 pKa = 4.71GDD218 pKa = 3.79PTDD221 pKa = 4.14DD222 pKa = 3.78QGHH225 pKa = 5.73GTHH228 pKa = 7.07CSGTIAAAANGIGVAGVAPNVNVMPLKK255 pKa = 10.68FLGSDD260 pKa = 3.59GSGSFSDD267 pKa = 3.43AVLAIEE273 pKa = 4.24YY274 pKa = 10.03AEE276 pKa = 4.62AMGADD281 pKa = 4.37VISASWGGYY290 pKa = 7.71TIGVDD295 pKa = 3.04SSMYY299 pKa = 10.55DD300 pKa = 3.51AINDD304 pKa = 3.71FSGLFCAAAGNDD316 pKa = 4.22GYY318 pKa = 11.64DD319 pKa = 3.12MDD321 pKa = 4.53FYY323 pKa = 11.81GMFGYY328 pKa = 9.92HH329 pKa = 6.76HH330 pKa = 7.44APATYY335 pKa = 8.91PNEE338 pKa = 4.43NIVSVASIDD347 pKa = 3.69SDD349 pKa = 3.7GSLSYY354 pKa = 10.35FSNYY358 pKa = 8.78GRR360 pKa = 11.84TSVDD364 pKa = 3.12LVSPGRR370 pKa = 11.84SILSTVPGDD379 pKa = 3.53SYY381 pKa = 11.44GYY383 pKa = 10.82KK384 pKa = 10.1SGTSMATPHH393 pKa = 5.82VAGTVALMISHH404 pKa = 7.75DD405 pKa = 3.49IDD407 pKa = 4.61LNGSVTRR414 pKa = 11.84SKK416 pKa = 11.17VQLKK420 pKa = 10.64DD421 pKa = 3.64DD422 pKa = 4.08LLEE425 pKa = 4.16ATVYY429 pKa = 10.6KK430 pKa = 9.61STYY433 pKa = 9.73NSVVLDD439 pKa = 3.89GCLNAYY445 pKa = 7.82EE446 pKa = 4.75ALLLTTDD453 pKa = 4.51NGTEE457 pKa = 4.04PPPPPEE463 pKa = 5.31NEE465 pKa = 4.26DD466 pKa = 3.88PYY468 pKa = 11.4NDD470 pKa = 3.75TDD472 pKa = 3.99FPPYY476 pKa = 8.19MTGTFEE482 pKa = 4.07VGGAIIAHH490 pKa = 6.25TGTWIDD496 pKa = 4.29EE497 pKa = 4.23GDD499 pKa = 3.56LDD501 pKa = 5.74YY502 pKa = 11.75YY503 pKa = 9.11CTWQVTADD511 pKa = 3.98PEE513 pKa = 4.22EE514 pKa = 5.03DD515 pKa = 3.59SPSDD519 pKa = 3.56YY520 pKa = 11.31EE521 pKa = 3.92EE522 pKa = 4.82WYY524 pKa = 10.48DD525 pKa = 4.23EE526 pKa = 3.94SWLYY530 pKa = 11.27EE531 pKa = 4.16EE532 pKa = 5.38FPPIFIVEE540 pKa = 4.38PGQAEE545 pKa = 4.18KK546 pKa = 10.81YY547 pKa = 10.03IRR549 pKa = 11.84IEE551 pKa = 3.96VVAVDD556 pKa = 3.91EE557 pKa = 4.73EE558 pKa = 4.63GLDD561 pKa = 3.72GVAYY565 pKa = 10.54SDD567 pKa = 3.86WEE569 pKa = 4.39WIEE572 pKa = 3.98PEE574 pKa = 4.57APNEE578 pKa = 3.97PPVNDD583 pKa = 3.49GDD585 pKa = 4.06RR586 pKa = 11.84PSVTGLMIAGEE597 pKa = 4.3TVYY600 pKa = 11.05AEE602 pKa = 4.31DD603 pKa = 5.39GGWSDD608 pKa = 5.07DD609 pKa = 3.71GGVEE613 pKa = 4.01NLEE616 pKa = 3.96YY617 pKa = 10.86SYY619 pKa = 10.05MWQISDD625 pKa = 4.12TEE627 pKa = 4.26FGTYY631 pKa = 10.12EE632 pKa = 4.15DD633 pKa = 3.94TGVTADD639 pKa = 3.95SYY641 pKa = 11.63SLTEE645 pKa = 4.0AEE647 pKa = 4.04VTKK650 pKa = 9.9FIRR653 pKa = 11.84VLVTAIDD660 pKa = 3.63NGDD663 pKa = 3.82PVEE666 pKa = 4.01MSEE669 pKa = 4.17PSEE672 pKa = 4.34SPGRR676 pKa = 11.84QVIPNAAPVNDD687 pKa = 3.81ITPSITGTLIAGDD700 pKa = 3.53IAYY703 pKa = 10.24ADD705 pKa = 3.49IGDD708 pKa = 3.72WSDD711 pKa = 4.69DD712 pKa = 3.49RR713 pKa = 11.84GEE715 pKa = 4.28EE716 pKa = 4.02NLSYY720 pKa = 10.82SYY722 pKa = 10.79NWKK725 pKa = 10.12IKK727 pKa = 9.98EE728 pKa = 4.01LDD730 pKa = 3.35GSYY733 pKa = 11.09SDD735 pKa = 5.31IGGSDD740 pKa = 3.64NPLSLPEE747 pKa = 4.84SDD749 pKa = 2.51TGYY752 pKa = 10.39YY753 pKa = 9.66IKK755 pKa = 11.09LEE757 pKa = 4.0VTATDD762 pKa = 3.46KK763 pKa = 11.39GDD765 pKa = 3.71PVKK768 pKa = 9.17WTSEE772 pKa = 3.72SSADD776 pKa = 3.79YY777 pKa = 10.47LVEE780 pKa = 4.06PAEE783 pKa = 4.33TPVINTFTSSITDD796 pKa = 4.08AYY798 pKa = 8.01PTSIRR803 pKa = 11.84NKK805 pKa = 6.99WTATVTVSTNLPMSEE820 pKa = 4.66AEE822 pKa = 4.67VAYY825 pKa = 9.44EE826 pKa = 3.77WSDD829 pKa = 3.54GKK831 pKa = 11.21SVTAGTAPTINGEE844 pKa = 4.35CTVISDD850 pKa = 5.48PIRR853 pKa = 11.84KK854 pKa = 9.35NIGSITFTIIGITPLDD870 pKa = 3.85NDD872 pKa = 3.29SDD874 pKa = 4.38YY875 pKa = 11.49TFDD878 pKa = 4.48PAHH881 pKa = 7.03SDD883 pKa = 3.23LTVTVNKK890 pKa = 9.87PP891 pKa = 2.85
MM1 pKa = 7.19KK2 pKa = 9.94RR3 pKa = 11.84KK4 pKa = 9.23SRR6 pKa = 11.84AVIYY10 pKa = 10.35VILAVILLLSTFVRR24 pKa = 11.84AYY26 pKa = 10.72GDD28 pKa = 3.54YY29 pKa = 9.97GTGNLSSDD37 pKa = 3.58GQQEE41 pKa = 4.38KK42 pKa = 10.6IGITEE47 pKa = 4.01YY48 pKa = 11.3LNDD51 pKa = 3.64QIIVKK56 pKa = 10.17FKK58 pKa = 11.23DD59 pKa = 4.2DD60 pKa = 4.39ISDD63 pKa = 3.58TDD65 pKa = 3.21RR66 pKa = 11.84KK67 pKa = 10.35LIKK70 pKa = 10.01KK71 pKa = 9.4YY72 pKa = 11.02YY73 pKa = 10.28KK74 pKa = 10.7LIDD77 pKa = 3.35KK78 pKa = 10.88AKK80 pKa = 8.51FKK82 pKa = 11.22SGGEE86 pKa = 3.99VLKK89 pKa = 11.2LKK91 pKa = 10.98GNLTVEE97 pKa = 4.66EE98 pKa = 4.22LVEE101 pKa = 3.96MLGEE105 pKa = 4.28LDD107 pKa = 3.65SVEE110 pKa = 4.24YY111 pKa = 10.96AGFDD115 pKa = 3.73YY116 pKa = 10.87IMRR119 pKa = 11.84ATDD122 pKa = 4.03FPDD125 pKa = 4.5DD126 pKa = 3.82PWDD129 pKa = 4.16GDD131 pKa = 3.79SARR134 pKa = 11.84YY135 pKa = 8.16LWGMEE140 pKa = 4.32KK141 pKa = 9.27IDD143 pKa = 3.97ANGDD147 pKa = 3.54TEE149 pKa = 6.03YY150 pKa = 10.78PGAWNLATGNPEE162 pKa = 3.99VIVAVIDD169 pKa = 3.46TGVDD173 pKa = 3.06MGHH176 pKa = 6.49VDD178 pKa = 3.68LAEE181 pKa = 4.56NIWINEE187 pKa = 3.86VEE189 pKa = 4.3LNGDD193 pKa = 3.81AGVDD197 pKa = 3.67DD198 pKa = 5.66DD199 pKa = 5.1GNGYY203 pKa = 9.68IDD205 pKa = 6.01DD206 pKa = 3.63VHH208 pKa = 7.62GWDD211 pKa = 3.81FHH213 pKa = 8.45NGDD216 pKa = 4.71GDD218 pKa = 3.79PTDD221 pKa = 4.14DD222 pKa = 3.78QGHH225 pKa = 5.73GTHH228 pKa = 7.07CSGTIAAAANGIGVAGVAPNVNVMPLKK255 pKa = 10.68FLGSDD260 pKa = 3.59GSGSFSDD267 pKa = 3.43AVLAIEE273 pKa = 4.24YY274 pKa = 10.03AEE276 pKa = 4.62AMGADD281 pKa = 4.37VISASWGGYY290 pKa = 7.71TIGVDD295 pKa = 3.04SSMYY299 pKa = 10.55DD300 pKa = 3.51AINDD304 pKa = 3.71FSGLFCAAAGNDD316 pKa = 4.22GYY318 pKa = 11.64DD319 pKa = 3.12MDD321 pKa = 4.53FYY323 pKa = 11.81GMFGYY328 pKa = 9.92HH329 pKa = 6.76HH330 pKa = 7.44APATYY335 pKa = 8.91PNEE338 pKa = 4.43NIVSVASIDD347 pKa = 3.69SDD349 pKa = 3.7GSLSYY354 pKa = 10.35FSNYY358 pKa = 8.78GRR360 pKa = 11.84TSVDD364 pKa = 3.12LVSPGRR370 pKa = 11.84SILSTVPGDD379 pKa = 3.53SYY381 pKa = 11.44GYY383 pKa = 10.82KK384 pKa = 10.1SGTSMATPHH393 pKa = 5.82VAGTVALMISHH404 pKa = 7.75DD405 pKa = 3.49IDD407 pKa = 4.61LNGSVTRR414 pKa = 11.84SKK416 pKa = 11.17VQLKK420 pKa = 10.64DD421 pKa = 3.64DD422 pKa = 4.08LLEE425 pKa = 4.16ATVYY429 pKa = 10.6KK430 pKa = 9.61STYY433 pKa = 9.73NSVVLDD439 pKa = 3.89GCLNAYY445 pKa = 7.82EE446 pKa = 4.75ALLLTTDD453 pKa = 4.51NGTEE457 pKa = 4.04PPPPPEE463 pKa = 5.31NEE465 pKa = 4.26DD466 pKa = 3.88PYY468 pKa = 11.4NDD470 pKa = 3.75TDD472 pKa = 3.99FPPYY476 pKa = 8.19MTGTFEE482 pKa = 4.07VGGAIIAHH490 pKa = 6.25TGTWIDD496 pKa = 4.29EE497 pKa = 4.23GDD499 pKa = 3.56LDD501 pKa = 5.74YY502 pKa = 11.75YY503 pKa = 9.11CTWQVTADD511 pKa = 3.98PEE513 pKa = 4.22EE514 pKa = 5.03DD515 pKa = 3.59SPSDD519 pKa = 3.56YY520 pKa = 11.31EE521 pKa = 3.92EE522 pKa = 4.82WYY524 pKa = 10.48DD525 pKa = 4.23EE526 pKa = 3.94SWLYY530 pKa = 11.27EE531 pKa = 4.16EE532 pKa = 5.38FPPIFIVEE540 pKa = 4.38PGQAEE545 pKa = 4.18KK546 pKa = 10.81YY547 pKa = 10.03IRR549 pKa = 11.84IEE551 pKa = 3.96VVAVDD556 pKa = 3.91EE557 pKa = 4.73EE558 pKa = 4.63GLDD561 pKa = 3.72GVAYY565 pKa = 10.54SDD567 pKa = 3.86WEE569 pKa = 4.39WIEE572 pKa = 3.98PEE574 pKa = 4.57APNEE578 pKa = 3.97PPVNDD583 pKa = 3.49GDD585 pKa = 4.06RR586 pKa = 11.84PSVTGLMIAGEE597 pKa = 4.3TVYY600 pKa = 11.05AEE602 pKa = 4.31DD603 pKa = 5.39GGWSDD608 pKa = 5.07DD609 pKa = 3.71GGVEE613 pKa = 4.01NLEE616 pKa = 3.96YY617 pKa = 10.86SYY619 pKa = 10.05MWQISDD625 pKa = 4.12TEE627 pKa = 4.26FGTYY631 pKa = 10.12EE632 pKa = 4.15DD633 pKa = 3.94TGVTADD639 pKa = 3.95SYY641 pKa = 11.63SLTEE645 pKa = 4.0AEE647 pKa = 4.04VTKK650 pKa = 9.9FIRR653 pKa = 11.84VLVTAIDD660 pKa = 3.63NGDD663 pKa = 3.82PVEE666 pKa = 4.01MSEE669 pKa = 4.17PSEE672 pKa = 4.34SPGRR676 pKa = 11.84QVIPNAAPVNDD687 pKa = 3.81ITPSITGTLIAGDD700 pKa = 3.53IAYY703 pKa = 10.24ADD705 pKa = 3.49IGDD708 pKa = 3.72WSDD711 pKa = 4.69DD712 pKa = 3.49RR713 pKa = 11.84GEE715 pKa = 4.28EE716 pKa = 4.02NLSYY720 pKa = 10.82SYY722 pKa = 10.79NWKK725 pKa = 10.12IKK727 pKa = 9.98EE728 pKa = 4.01LDD730 pKa = 3.35GSYY733 pKa = 11.09SDD735 pKa = 5.31IGGSDD740 pKa = 3.64NPLSLPEE747 pKa = 4.84SDD749 pKa = 2.51TGYY752 pKa = 10.39YY753 pKa = 9.66IKK755 pKa = 11.09LEE757 pKa = 4.0VTATDD762 pKa = 3.46KK763 pKa = 11.39GDD765 pKa = 3.71PVKK768 pKa = 9.17WTSEE772 pKa = 3.72SSADD776 pKa = 3.79YY777 pKa = 10.47LVEE780 pKa = 4.06PAEE783 pKa = 4.33TPVINTFTSSITDD796 pKa = 4.08AYY798 pKa = 8.01PTSIRR803 pKa = 11.84NKK805 pKa = 6.99WTATVTVSTNLPMSEE820 pKa = 4.66AEE822 pKa = 4.67VAYY825 pKa = 9.44EE826 pKa = 3.77WSDD829 pKa = 3.54GKK831 pKa = 11.21SVTAGTAPTINGEE844 pKa = 4.35CTVISDD850 pKa = 5.48PIRR853 pKa = 11.84KK854 pKa = 9.35NIGSITFTIIGITPLDD870 pKa = 3.85NDD872 pKa = 3.29SDD874 pKa = 4.38YY875 pKa = 11.49TFDD878 pKa = 4.48PAHH881 pKa = 7.03SDD883 pKa = 3.23LTVTVNKK890 pKa = 9.87PP891 pKa = 2.85
Molecular weight: 96.59 kDa
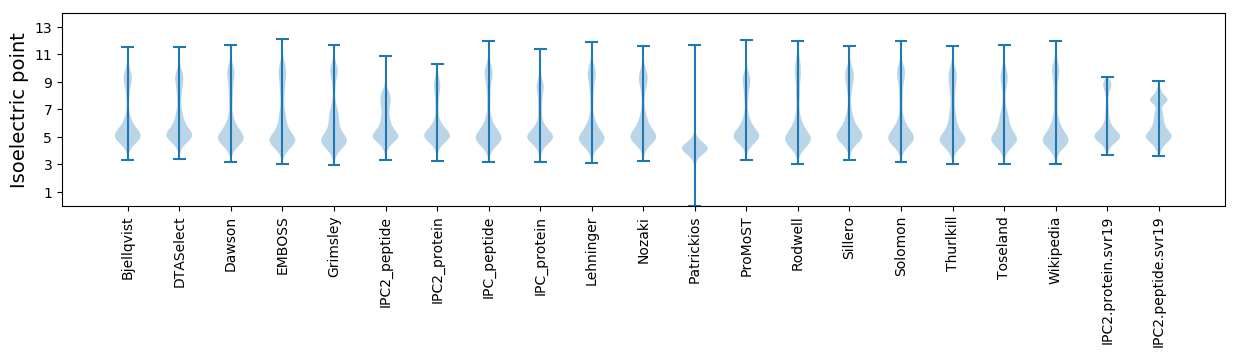
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6LG06|A0A1M6LG06_9FIRM Uncharacterized protein OS=Dethiosulfatibacter aminovorans DSM 17477 OX=1121476 GN=SAMN02745751_03162 PE=4 SV=1
MM1 pKa = 7.83AKK3 pKa = 9.7TSMKK7 pKa = 10.25VKK9 pKa = 9.39QQRR12 pKa = 11.84KK13 pKa = 6.65QKK15 pKa = 10.28FSTRR19 pKa = 11.84EE20 pKa = 3.53YY21 pKa = 10.22SRR23 pKa = 11.84CRR25 pKa = 11.84ICGRR29 pKa = 11.84PHH31 pKa = 7.1AYY33 pKa = 9.15LRR35 pKa = 11.84KK36 pKa = 9.67YY37 pKa = 9.37GICRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.97LAYY49 pKa = 9.84KK50 pKa = 10.14GQIPGVRR57 pKa = 11.84KK58 pKa = 10.13ASWW61 pKa = 2.78
MM1 pKa = 7.83AKK3 pKa = 9.7TSMKK7 pKa = 10.25VKK9 pKa = 9.39QQRR12 pKa = 11.84KK13 pKa = 6.65QKK15 pKa = 10.28FSTRR19 pKa = 11.84EE20 pKa = 3.53YY21 pKa = 10.22SRR23 pKa = 11.84CRR25 pKa = 11.84ICGRR29 pKa = 11.84PHH31 pKa = 7.1AYY33 pKa = 9.15LRR35 pKa = 11.84KK36 pKa = 9.67YY37 pKa = 9.37GICRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.97LAYY49 pKa = 9.84KK50 pKa = 10.14GQIPGVRR57 pKa = 11.84KK58 pKa = 10.13ASWW61 pKa = 2.78
Molecular weight: 7.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1182095 |
39 |
2390 |
325.8 |
36.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.475 ± 0.047 | 1.168 ± 0.015 |
5.982 ± 0.036 | 7.826 ± 0.046 |
4.304 ± 0.03 | 7.294 ± 0.041 |
1.529 ± 0.014 | 8.98 ± 0.041 |
7.415 ± 0.043 | 8.764 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.286 ± 0.022 | 4.977 ± 0.035 |
3.084 ± 0.022 | 2.124 ± 0.017 |
3.951 ± 0.026 | 6.181 ± 0.03 |
4.955 ± 0.03 | 7.05 ± 0.032 |
0.709 ± 0.013 | 3.945 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
