
Desulfovibrio marinus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfovibrionales; Desulfovibrionaceae; Desulfovibrio
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
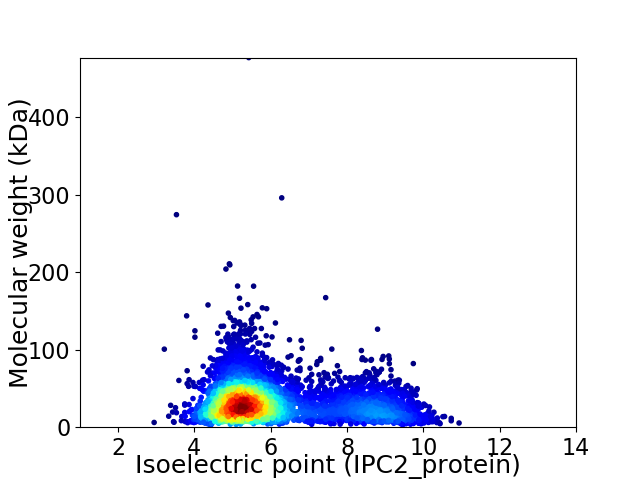
Virtual 2D-PAGE plot for 4594 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P1ZGT0|A0A6P1ZGT0_9DELT Phenylacetate-coenzyme A ligase OS=Desulfovibrio marinus OX=370038 GN=DQK91_10020 PE=3 SV=1
MM1 pKa = 6.99STLTGAMYY9 pKa = 9.82TGIAGLTTHH18 pKa = 6.57SKK20 pKa = 10.54GISVAGNNLANSSTVGYY37 pKa = 9.79KK38 pKa = 8.89STSIQFEE45 pKa = 4.48DD46 pKa = 3.27MFYY49 pKa = 11.23SSLHH53 pKa = 5.36TANGPDD59 pKa = 3.72QIGHH63 pKa = 6.43GSSVSTLYY71 pKa = 11.1SDD73 pKa = 5.3FSAGSYY79 pKa = 10.73EE80 pKa = 4.34DD81 pKa = 3.66TASPTDD87 pKa = 3.59VACTGKK93 pKa = 10.77GFFVVTDD100 pKa = 3.63PTTGRR105 pKa = 11.84NYY107 pKa = 8.14YY108 pKa = 8.77TRR110 pKa = 11.84AGHH113 pKa = 6.93FSFNEE118 pKa = 3.64DD119 pKa = 3.13GYY121 pKa = 11.29LINAQGYY128 pKa = 8.3RR129 pKa = 11.84VQGWEE134 pKa = 4.32ADD136 pKa = 3.76PAHH139 pKa = 7.05KK140 pKa = 10.91GNGVSTKK147 pKa = 10.54GALGDD152 pKa = 3.66IRR154 pKa = 11.84FDD156 pKa = 4.14DD157 pKa = 4.33LQSPPEE163 pKa = 4.08ATSFMTLLVNLDD175 pKa = 3.45KK176 pKa = 11.32DD177 pKa = 3.87SEE179 pKa = 4.21EE180 pKa = 4.5RR181 pKa = 11.84SANPTDD187 pKa = 3.98PFFSLQQEE195 pKa = 4.23WDD197 pKa = 3.57GTADD201 pKa = 3.43PPMGEE206 pKa = 3.75NSYY209 pKa = 9.36TYY211 pKa = 9.12QSSMVVYY218 pKa = 10.28DD219 pKa = 3.78EE220 pKa = 5.54AGGSHH225 pKa = 6.41EE226 pKa = 4.02VTIYY230 pKa = 10.59FDD232 pKa = 3.79PVQDD236 pKa = 4.11DD237 pKa = 4.36SVVSDD242 pKa = 3.6AGGGQVWEE250 pKa = 4.57YY251 pKa = 10.6IITCDD256 pKa = 3.47PSEE259 pKa = 4.81DD260 pKa = 3.28GRR262 pKa = 11.84TIDD265 pKa = 3.77GQEE268 pKa = 3.92LSSTSSAGLLMTGTLTFDD286 pKa = 4.04SSSQLTGMTAFTLNGAATGDD306 pKa = 3.86LHH308 pKa = 7.5DD309 pKa = 4.42LTNWTPADD317 pKa = 3.78IGDD320 pKa = 4.85DD321 pKa = 4.01GNPMFTANFSGSDD334 pKa = 3.35NASYY338 pKa = 10.08TGDD341 pKa = 3.75AEE343 pKa = 4.23ALNIGIDD350 pKa = 4.28FGIHH354 pKa = 7.2DD355 pKa = 4.81NTPSATGWDD364 pKa = 3.7SAVADD369 pKa = 4.17ASLIGSDD376 pKa = 3.66IANLFNFEE384 pKa = 4.17EE385 pKa = 5.14PIISSNATTAYY396 pKa = 9.77GRR398 pKa = 11.84SSSTLSRR405 pKa = 11.84SQDD408 pKa = 3.04GYY410 pKa = 10.45PPGFLEE416 pKa = 5.79DD417 pKa = 4.89VEE419 pKa = 5.36IDD421 pKa = 3.74DD422 pKa = 4.65NGVITGIFSNDD433 pKa = 2.93QSMEE437 pKa = 4.18LYY439 pKa = 10.86VLGLADD445 pKa = 4.14FANYY449 pKa = 9.59QGLINEE455 pKa = 4.75GGNLYY460 pKa = 10.4SASGDD465 pKa = 3.27SGLPITGVAGTNQFGSVASNKK486 pKa = 10.19LEE488 pKa = 4.04LSNVDD493 pKa = 3.37YY494 pKa = 10.97SGEE497 pKa = 4.05MVDD500 pKa = 4.65LIRR503 pKa = 11.84YY504 pKa = 6.69QRR506 pKa = 11.84GYY508 pKa = 10.36QSNSKK513 pKa = 10.68VITTVDD519 pKa = 3.32TLLQEE524 pKa = 4.98AINLKK529 pKa = 10.3RR530 pKa = 3.64
MM1 pKa = 6.99STLTGAMYY9 pKa = 9.82TGIAGLTTHH18 pKa = 6.57SKK20 pKa = 10.54GISVAGNNLANSSTVGYY37 pKa = 9.79KK38 pKa = 8.89STSIQFEE45 pKa = 4.48DD46 pKa = 3.27MFYY49 pKa = 11.23SSLHH53 pKa = 5.36TANGPDD59 pKa = 3.72QIGHH63 pKa = 6.43GSSVSTLYY71 pKa = 11.1SDD73 pKa = 5.3FSAGSYY79 pKa = 10.73EE80 pKa = 4.34DD81 pKa = 3.66TASPTDD87 pKa = 3.59VACTGKK93 pKa = 10.77GFFVVTDD100 pKa = 3.63PTTGRR105 pKa = 11.84NYY107 pKa = 8.14YY108 pKa = 8.77TRR110 pKa = 11.84AGHH113 pKa = 6.93FSFNEE118 pKa = 3.64DD119 pKa = 3.13GYY121 pKa = 11.29LINAQGYY128 pKa = 8.3RR129 pKa = 11.84VQGWEE134 pKa = 4.32ADD136 pKa = 3.76PAHH139 pKa = 7.05KK140 pKa = 10.91GNGVSTKK147 pKa = 10.54GALGDD152 pKa = 3.66IRR154 pKa = 11.84FDD156 pKa = 4.14DD157 pKa = 4.33LQSPPEE163 pKa = 4.08ATSFMTLLVNLDD175 pKa = 3.45KK176 pKa = 11.32DD177 pKa = 3.87SEE179 pKa = 4.21EE180 pKa = 4.5RR181 pKa = 11.84SANPTDD187 pKa = 3.98PFFSLQQEE195 pKa = 4.23WDD197 pKa = 3.57GTADD201 pKa = 3.43PPMGEE206 pKa = 3.75NSYY209 pKa = 9.36TYY211 pKa = 9.12QSSMVVYY218 pKa = 10.28DD219 pKa = 3.78EE220 pKa = 5.54AGGSHH225 pKa = 6.41EE226 pKa = 4.02VTIYY230 pKa = 10.59FDD232 pKa = 3.79PVQDD236 pKa = 4.11DD237 pKa = 4.36SVVSDD242 pKa = 3.6AGGGQVWEE250 pKa = 4.57YY251 pKa = 10.6IITCDD256 pKa = 3.47PSEE259 pKa = 4.81DD260 pKa = 3.28GRR262 pKa = 11.84TIDD265 pKa = 3.77GQEE268 pKa = 3.92LSSTSSAGLLMTGTLTFDD286 pKa = 4.04SSSQLTGMTAFTLNGAATGDD306 pKa = 3.86LHH308 pKa = 7.5DD309 pKa = 4.42LTNWTPADD317 pKa = 3.78IGDD320 pKa = 4.85DD321 pKa = 4.01GNPMFTANFSGSDD334 pKa = 3.35NASYY338 pKa = 10.08TGDD341 pKa = 3.75AEE343 pKa = 4.23ALNIGIDD350 pKa = 4.28FGIHH354 pKa = 7.2DD355 pKa = 4.81NTPSATGWDD364 pKa = 3.7SAVADD369 pKa = 4.17ASLIGSDD376 pKa = 3.66IANLFNFEE384 pKa = 4.17EE385 pKa = 5.14PIISSNATTAYY396 pKa = 9.77GRR398 pKa = 11.84SSSTLSRR405 pKa = 11.84SQDD408 pKa = 3.04GYY410 pKa = 10.45PPGFLEE416 pKa = 5.79DD417 pKa = 4.89VEE419 pKa = 5.36IDD421 pKa = 3.74DD422 pKa = 4.65NGVITGIFSNDD433 pKa = 2.93QSMEE437 pKa = 4.18LYY439 pKa = 10.86VLGLADD445 pKa = 4.14FANYY449 pKa = 9.59QGLINEE455 pKa = 4.75GGNLYY460 pKa = 10.4SASGDD465 pKa = 3.27SGLPITGVAGTNQFGSVASNKK486 pKa = 10.19LEE488 pKa = 4.04LSNVDD493 pKa = 3.37YY494 pKa = 10.97SGEE497 pKa = 4.05MVDD500 pKa = 4.65LIRR503 pKa = 11.84YY504 pKa = 6.69QRR506 pKa = 11.84GYY508 pKa = 10.36QSNSKK513 pKa = 10.68VITTVDD519 pKa = 3.32TLLQEE524 pKa = 4.98AINLKK529 pKa = 10.3RR530 pKa = 3.64
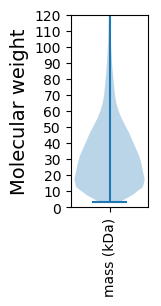
Molecular weight: 56.2 kDa
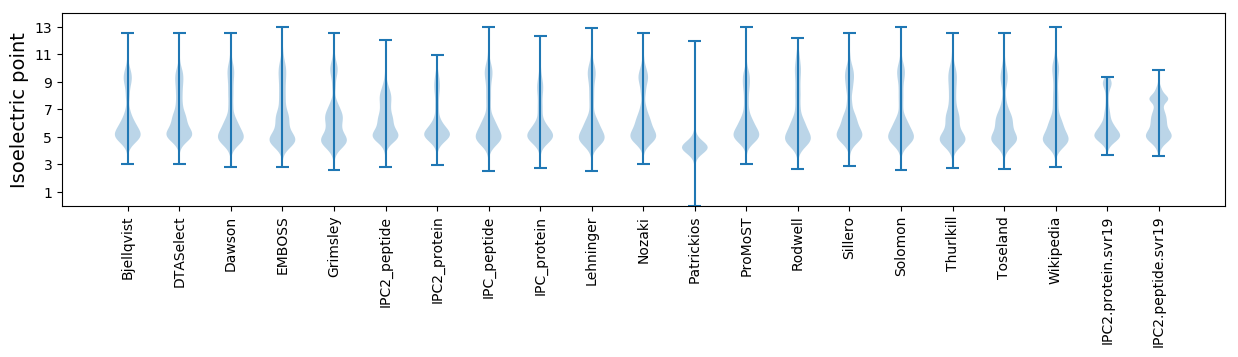
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P1ZMQ3|A0A6P1ZMQ3_9DELT Carbon monoxide dehydrogenase OS=Desulfovibrio marinus OX=370038 GN=DQK91_02450 PE=4 SV=1
MM1 pKa = 7.75SKK3 pKa = 10.38RR4 pKa = 11.84PYY6 pKa = 9.63QPSKK10 pKa = 9.73IRR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 8.13RR15 pKa = 11.84THH17 pKa = 5.96GFLVRR22 pKa = 11.84SRR24 pKa = 11.84TKK26 pKa = 10.05SGRR29 pKa = 11.84AVLRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.59GRR40 pKa = 11.84KK41 pKa = 8.88RR42 pKa = 11.84LACC45 pKa = 4.34
MM1 pKa = 7.75SKK3 pKa = 10.38RR4 pKa = 11.84PYY6 pKa = 9.63QPSKK10 pKa = 9.73IRR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 8.13RR15 pKa = 11.84THH17 pKa = 5.96GFLVRR22 pKa = 11.84SRR24 pKa = 11.84TKK26 pKa = 10.05SGRR29 pKa = 11.84AVLRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.59GRR40 pKa = 11.84KK41 pKa = 8.88RR42 pKa = 11.84LACC45 pKa = 4.34
Molecular weight: 5.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1459414 |
27 |
4366 |
317.7 |
34.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.376 ± 0.044 | 1.246 ± 0.019 |
5.702 ± 0.03 | 6.785 ± 0.039 |
3.884 ± 0.024 | 7.903 ± 0.036 |
2.169 ± 0.017 | 5.174 ± 0.03 |
4.065 ± 0.032 | 10.397 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.788 ± 0.016 | 2.904 ± 0.02 |
4.952 ± 0.028 | 3.308 ± 0.019 |
6.391 ± 0.037 | 5.679 ± 0.026 |
5.118 ± 0.022 | 7.312 ± 0.028 |
1.171 ± 0.014 | 2.678 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
