
Cryptosporangium arvum DSM 44712
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Cryptosporangiales; Cryptosporangiaceae; Cryptosporangium; Cryptosporangium arvum
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
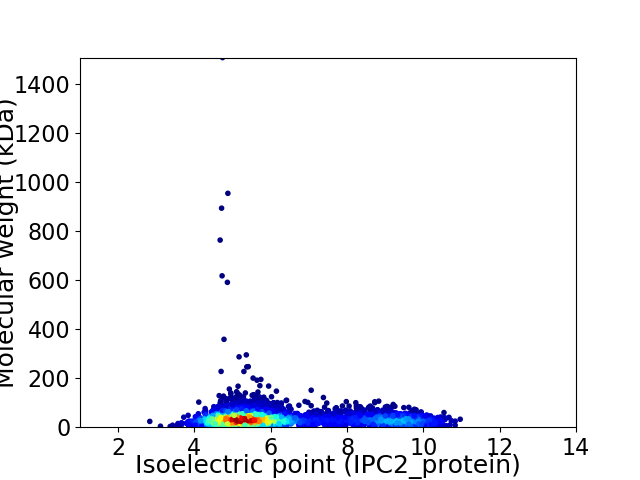
Virtual 2D-PAGE plot for 4033 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A010ZSE2|A0A010ZSE2_9ACTN Carbohydrate ABC transporter membrane protein 1 CUT1 family OS=Cryptosporangium arvum DSM 44712 OX=927661 GN=CryarDRAFT_2727 PE=3 SV=1
MM1 pKa = 7.12SALVKK6 pKa = 10.38RR7 pKa = 11.84LAAAGSLIAAGAALVLVPSPAYY29 pKa = 10.46ADD31 pKa = 3.65TVTVNYY37 pKa = 10.23SCEE40 pKa = 3.95IPLSGTQTGDD50 pKa = 3.32VEE52 pKa = 4.64VTITAPATATVGEE65 pKa = 4.57TVDD68 pKa = 3.52VTVTTGPIPILPPIDD83 pKa = 4.79LDD85 pKa = 4.09AGSVTPSGEE94 pKa = 4.16VTVSGAQTGAVAVTGTPNPEE114 pKa = 4.26PYY116 pKa = 9.66PANQPVQLTALTGQLTLTTAGEE138 pKa = 4.4VQLSPGAVNATATTIFGTFTIPCVPTATPTPVAAVISVSS177 pKa = 3.15
MM1 pKa = 7.12SALVKK6 pKa = 10.38RR7 pKa = 11.84LAAAGSLIAAGAALVLVPSPAYY29 pKa = 10.46ADD31 pKa = 3.65TVTVNYY37 pKa = 10.23SCEE40 pKa = 3.95IPLSGTQTGDD50 pKa = 3.32VEE52 pKa = 4.64VTITAPATATVGEE65 pKa = 4.57TVDD68 pKa = 3.52VTVTTGPIPILPPIDD83 pKa = 4.79LDD85 pKa = 4.09AGSVTPSGEE94 pKa = 4.16VTVSGAQTGAVAVTGTPNPEE114 pKa = 4.26PYY116 pKa = 9.66PANQPVQLTALTGQLTLTTAGEE138 pKa = 4.4VQLSPGAVNATATTIFGTFTIPCVPTATPTPVAAVISVSS177 pKa = 3.15
Molecular weight: 17.31 kDa
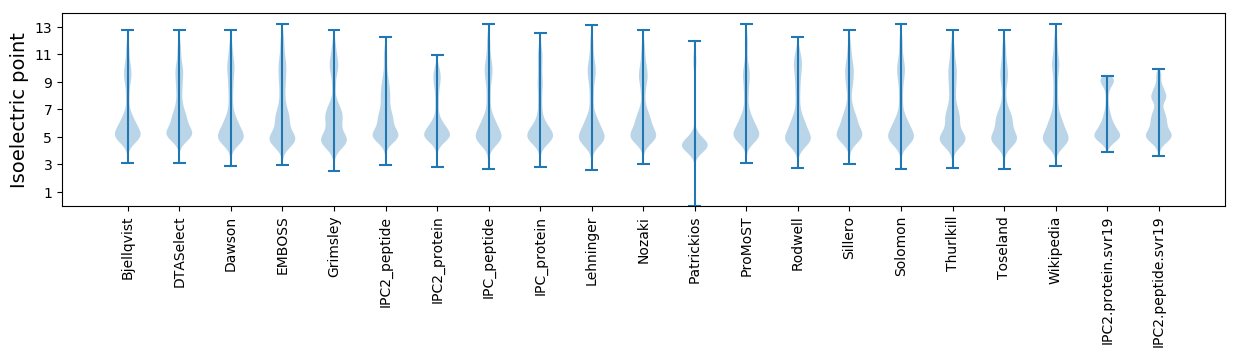
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A010ZQ45|A0A010ZQ45_9ACTN Kef-type K+ ransport system predicted NAD-binding component OS=Cryptosporangium arvum DSM 44712 OX=927661 GN=CryarDRAFT_1859 PE=4 SV=1
MM1 pKa = 7.21VSTVRR6 pKa = 11.84KK7 pKa = 8.51IRR9 pKa = 11.84TVRR12 pKa = 11.84TVSDD16 pKa = 3.24VHH18 pKa = 4.84TVRR21 pKa = 11.84NIRR24 pKa = 11.84TSRR27 pKa = 11.84SVCTMSTVRR36 pKa = 11.84ISRR39 pKa = 11.84NVRR42 pKa = 11.84TSRR45 pKa = 11.84NVCTMSTVRR54 pKa = 11.84TSRR57 pKa = 11.84NVRR60 pKa = 11.84TMSTVRR66 pKa = 11.84TSHH69 pKa = 5.87NVRR72 pKa = 11.84TMSTVRR78 pKa = 11.84TTRR81 pKa = 11.84NVRR84 pKa = 11.84TMRR87 pKa = 11.84ALRR90 pKa = 11.84FVRR93 pKa = 11.84PVRR96 pKa = 11.84LIPALRR102 pKa = 11.84HH103 pKa = 4.96VSTLRR108 pKa = 11.84HH109 pKa = 5.62VGTLHH114 pKa = 6.7FVRR117 pKa = 11.84INGKK121 pKa = 9.22RR122 pKa = 11.84LSEE125 pKa = 4.0RR126 pKa = 11.84DD127 pKa = 3.1AQRR130 pKa = 11.84ILRR133 pKa = 11.84RR134 pKa = 11.84LHH136 pKa = 5.96RR137 pKa = 11.84RR138 pKa = 11.84RR139 pKa = 11.84SLRR142 pKa = 11.84RR143 pKa = 11.84RR144 pKa = 11.84AVLRR148 pKa = 11.84GTVQQRR154 pKa = 11.84TLRR157 pKa = 11.84RR158 pKa = 11.84PTLQRR163 pKa = 11.84RR164 pKa = 11.84TLRR167 pKa = 11.84RR168 pKa = 11.84RR169 pKa = 11.84SAHH172 pKa = 4.94EE173 pKa = 3.67LRR175 pKa = 11.84RR176 pKa = 11.84GKK178 pKa = 7.92QLRR181 pKa = 11.84RR182 pKa = 11.84SRR184 pKa = 11.84SRR186 pKa = 11.84LKK188 pKa = 10.54IGWIGHH194 pKa = 4.43TVRR197 pKa = 11.84LRR199 pKa = 11.84RR200 pKa = 11.84VVNKK204 pKa = 9.7PPAPISDD211 pKa = 3.81HH212 pKa = 5.67PRR214 pKa = 11.84SRR216 pKa = 11.84ILQCRR221 pKa = 11.84RR222 pKa = 11.84VLQRR226 pKa = 11.84GRR228 pKa = 11.84PGLHH232 pKa = 4.52VRR234 pKa = 11.84IRR236 pKa = 11.84RR237 pKa = 11.84QAEE240 pKa = 3.82IRR242 pKa = 11.84PGPPRR247 pKa = 11.84PLPPPIAVPVNEE259 pKa = 4.11QQRR262 pKa = 11.84PGHH265 pKa = 4.76SRR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84TLRR274 pKa = 3.46
MM1 pKa = 7.21VSTVRR6 pKa = 11.84KK7 pKa = 8.51IRR9 pKa = 11.84TVRR12 pKa = 11.84TVSDD16 pKa = 3.24VHH18 pKa = 4.84TVRR21 pKa = 11.84NIRR24 pKa = 11.84TSRR27 pKa = 11.84SVCTMSTVRR36 pKa = 11.84ISRR39 pKa = 11.84NVRR42 pKa = 11.84TSRR45 pKa = 11.84NVCTMSTVRR54 pKa = 11.84TSRR57 pKa = 11.84NVRR60 pKa = 11.84TMSTVRR66 pKa = 11.84TSHH69 pKa = 5.87NVRR72 pKa = 11.84TMSTVRR78 pKa = 11.84TTRR81 pKa = 11.84NVRR84 pKa = 11.84TMRR87 pKa = 11.84ALRR90 pKa = 11.84FVRR93 pKa = 11.84PVRR96 pKa = 11.84LIPALRR102 pKa = 11.84HH103 pKa = 4.96VSTLRR108 pKa = 11.84HH109 pKa = 5.62VGTLHH114 pKa = 6.7FVRR117 pKa = 11.84INGKK121 pKa = 9.22RR122 pKa = 11.84LSEE125 pKa = 4.0RR126 pKa = 11.84DD127 pKa = 3.1AQRR130 pKa = 11.84ILRR133 pKa = 11.84RR134 pKa = 11.84LHH136 pKa = 5.96RR137 pKa = 11.84RR138 pKa = 11.84RR139 pKa = 11.84SLRR142 pKa = 11.84RR143 pKa = 11.84RR144 pKa = 11.84AVLRR148 pKa = 11.84GTVQQRR154 pKa = 11.84TLRR157 pKa = 11.84RR158 pKa = 11.84PTLQRR163 pKa = 11.84RR164 pKa = 11.84TLRR167 pKa = 11.84RR168 pKa = 11.84RR169 pKa = 11.84SAHH172 pKa = 4.94EE173 pKa = 3.67LRR175 pKa = 11.84RR176 pKa = 11.84GKK178 pKa = 7.92QLRR181 pKa = 11.84RR182 pKa = 11.84SRR184 pKa = 11.84SRR186 pKa = 11.84LKK188 pKa = 10.54IGWIGHH194 pKa = 4.43TVRR197 pKa = 11.84LRR199 pKa = 11.84RR200 pKa = 11.84VVNKK204 pKa = 9.7PPAPISDD211 pKa = 3.81HH212 pKa = 5.67PRR214 pKa = 11.84SRR216 pKa = 11.84ILQCRR221 pKa = 11.84RR222 pKa = 11.84VLQRR226 pKa = 11.84GRR228 pKa = 11.84PGLHH232 pKa = 4.52VRR234 pKa = 11.84IRR236 pKa = 11.84RR237 pKa = 11.84QAEE240 pKa = 3.82IRR242 pKa = 11.84PGPPRR247 pKa = 11.84PLPPPIAVPVNEE259 pKa = 4.11QQRR262 pKa = 11.84PGHH265 pKa = 4.76SRR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84TLRR274 pKa = 3.46
Molecular weight: 32.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1372967 |
13 |
14508 |
340.4 |
36.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.16 ± 0.06 | 0.659 ± 0.01 |
6.159 ± 0.036 | 5.206 ± 0.03 |
2.767 ± 0.023 | 9.222 ± 0.05 |
2.059 ± 0.018 | 3.312 ± 0.036 |
1.613 ± 0.03 | 10.735 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.509 ± 0.016 | 1.715 ± 0.023 |
6.077 ± 0.043 | 2.487 ± 0.022 |
8.252 ± 0.047 | 5.2 ± 0.028 |
6.101 ± 0.032 | 9.342 ± 0.05 |
1.509 ± 0.017 | 1.915 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
