
Planktothricoides sp. SR001
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Oscillatoriales; Microcoleaceae; Planktothricoides; unclassified Planktothricoides
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
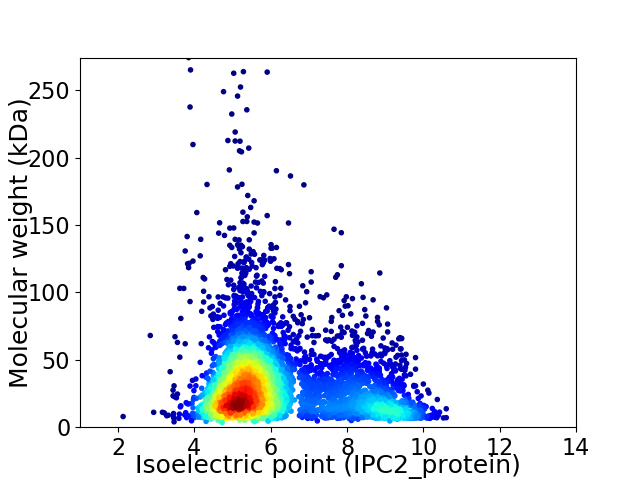
Virtual 2D-PAGE plot for 5042 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M1JMA9|A0A0M1JMA9_9CYAN Uncharacterized protein OS=Planktothricoides sp. SR001 OX=1705388 GN=AM228_27290 PE=4 SV=1
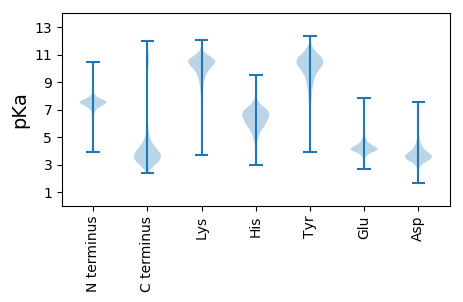
MM1 pKa = 7.71NYY3 pKa = 9.54QNKK6 pKa = 10.18AKK8 pKa = 9.76IQLISGVILASTTLTIPVNAQPITPAADD36 pKa = 3.53GTGTIVNQQDD46 pKa = 3.54NIINITEE53 pKa = 4.6GKK55 pKa = 9.91RR56 pKa = 11.84SSNGANLFHH65 pKa = 7.67SFDD68 pKa = 3.54QFGVNTGQTANFQSSPEE85 pKa = 3.83INNILGRR92 pKa = 11.84VVGGNASIINGILQVTGGNSNLFLMNPAGILFGPNASLNVPADD135 pKa = 4.05FTATTATSIGFGNNNWFQSIGDD157 pKa = 4.01NNWANLVGNPSDD169 pKa = 3.85FSFDD173 pKa = 3.32LAQPGWVVNLADD185 pKa = 3.59LTLNFGQNLTLLGGGILNSGNLSAPGGNISIAAVPGEE222 pKa = 4.09SLVRR226 pKa = 11.84ISQPGNILSLDD237 pKa = 3.21IATPGLNEE245 pKa = 4.04GVINPLSLPEE255 pKa = 4.41LLTGGSQILDD265 pKa = 3.4ATQVQHH271 pKa = 6.44NADD274 pKa = 3.41GTITLTSSGVTIDD287 pKa = 3.95PNTDD291 pKa = 2.81NGLALAAGDD300 pKa = 3.74VTTEE304 pKa = 3.53TGGQVNVLTGGGNIILDD321 pKa = 3.6QVNSDD326 pKa = 3.55KK327 pKa = 11.57VNINANGGNIAQVNSDD343 pKa = 3.65SLLNASAVQLQTYY356 pKa = 8.03GQGGIGLEE364 pKa = 4.23TEE366 pKa = 4.3PLRR369 pKa = 11.84LEE371 pKa = 4.43ANNLEE376 pKa = 4.2ATAGSGGAIFYY387 pKa = 10.26VPNGDD392 pKa = 3.51VTIGGVTDD400 pKa = 3.91EE401 pKa = 4.46LTGMSITDD409 pKa = 3.53GGDD412 pKa = 3.27FSLEE416 pKa = 4.17SVGSITLIEE425 pKa = 5.35DD426 pKa = 3.98IYY428 pKa = 11.14TSDD431 pKa = 5.37DD432 pKa = 3.52IQSGDD437 pKa = 3.03ITLTSNAGAIDD448 pKa = 3.78TTGGGISAYY457 pKa = 10.17SEE459 pKa = 4.17YY460 pKa = 11.29GNGGSVSLTAQGDD473 pKa = 3.49IHH475 pKa = 6.45TSNISSYY482 pKa = 10.99SPEE485 pKa = 3.75NGGNVSLEE493 pKa = 4.3STGGAIDD500 pKa = 3.61TTGGEE505 pKa = 3.84ISANSGYY512 pKa = 11.24GNGGSVSLTAQGDD525 pKa = 3.49IHH527 pKa = 6.48TSNISSDD534 pKa = 2.77SWFWTGGNVSLEE546 pKa = 4.31STNGAIDD553 pKa = 3.54TTGGEE558 pKa = 3.75ISANSVSGNAGDD570 pKa = 4.49VSLTAQGDD578 pKa = 3.47IHH580 pKa = 6.36TSNIRR585 pKa = 11.84SYY587 pKa = 11.09SSEE590 pKa = 3.91GNGGNVSLEE599 pKa = 4.27STGGEE604 pKa = 3.69IDD606 pKa = 3.55TTGGGILAGSGSSGGDD622 pKa = 2.99VSLIAQGDD630 pKa = 3.55IHH632 pKa = 6.45TSNIRR637 pKa = 11.84SFSWDD642 pKa = 3.06GTGGNVFLEE651 pKa = 4.52STGGAIDD658 pKa = 3.61TTGGEE663 pKa = 3.94ISPYY667 pKa = 9.72SAYY670 pKa = 10.69GNAGDD675 pKa = 4.44VSLIAQGDD683 pKa = 3.58IHH685 pKa = 6.27TSRR688 pKa = 11.84IGSYY692 pKa = 9.99SGEE695 pKa = 4.05GTGGNVFLEE704 pKa = 4.52STDD707 pKa = 3.63GAIDD711 pKa = 3.55TTGGEE716 pKa = 3.75ISANSVSGNGGSVSLTAQGDD736 pKa = 3.47IHH738 pKa = 6.36TSNIRR743 pKa = 11.84SYY745 pKa = 11.09SSEE748 pKa = 3.91GNGGNVSLEE757 pKa = 4.29STGGAINTSAGQLDD771 pKa = 3.93SHH773 pKa = 5.94SQEE776 pKa = 3.88GSAGEE781 pKa = 4.07IKK783 pKa = 9.92IDD785 pKa = 3.46ASGDD789 pKa = 3.23ILTGFVSSYY798 pKa = 9.68NQSSSADD805 pKa = 3.75SYY807 pKa = 12.0SGDD810 pKa = 2.67IWITSHH816 pKa = 6.5NGSINTTIGNLSGEE830 pKa = 4.33AQISPNDD837 pKa = 3.78DD838 pKa = 3.27VSSVEE843 pKa = 3.98VASIFNNAQANLASYY858 pKa = 9.68AHH860 pKa = 6.91SGTGGNVILKK870 pKa = 10.4ANGDD874 pKa = 3.34ITTSHH879 pKa = 7.1ISTFGSQSSGNVNIISNNGAINTGVIFSFSQLNNAGDD916 pKa = 3.75VTLNAAGNISTSHH929 pKa = 5.68ISAWGNQQGGNITIDD944 pKa = 3.22AGNIAGQINNNTPCDD959 pKa = 3.84CVNLLGTEE967 pKa = 4.08PTPSFNIGSATIQSFSQVGHH987 pKa = 7.03AGNVTITTSGDD998 pKa = 3.24TNLGGDD1004 pKa = 4.26EE1005 pKa = 4.04NRR1007 pKa = 11.84HH1008 pKa = 6.14AIRR1011 pKa = 11.84SAGYY1015 pKa = 8.06TEE1017 pKa = 5.08GGNISIFSLGG1027 pKa = 3.29
MM1 pKa = 7.71NYY3 pKa = 9.54QNKK6 pKa = 10.18AKK8 pKa = 9.76IQLISGVILASTTLTIPVNAQPITPAADD36 pKa = 3.53GTGTIVNQQDD46 pKa = 3.54NIINITEE53 pKa = 4.6GKK55 pKa = 9.91RR56 pKa = 11.84SSNGANLFHH65 pKa = 7.67SFDD68 pKa = 3.54QFGVNTGQTANFQSSPEE85 pKa = 3.83INNILGRR92 pKa = 11.84VVGGNASIINGILQVTGGNSNLFLMNPAGILFGPNASLNVPADD135 pKa = 4.05FTATTATSIGFGNNNWFQSIGDD157 pKa = 4.01NNWANLVGNPSDD169 pKa = 3.85FSFDD173 pKa = 3.32LAQPGWVVNLADD185 pKa = 3.59LTLNFGQNLTLLGGGILNSGNLSAPGGNISIAAVPGEE222 pKa = 4.09SLVRR226 pKa = 11.84ISQPGNILSLDD237 pKa = 3.21IATPGLNEE245 pKa = 4.04GVINPLSLPEE255 pKa = 4.41LLTGGSQILDD265 pKa = 3.4ATQVQHH271 pKa = 6.44NADD274 pKa = 3.41GTITLTSSGVTIDD287 pKa = 3.95PNTDD291 pKa = 2.81NGLALAAGDD300 pKa = 3.74VTTEE304 pKa = 3.53TGGQVNVLTGGGNIILDD321 pKa = 3.6QVNSDD326 pKa = 3.55KK327 pKa = 11.57VNINANGGNIAQVNSDD343 pKa = 3.65SLLNASAVQLQTYY356 pKa = 8.03GQGGIGLEE364 pKa = 4.23TEE366 pKa = 4.3PLRR369 pKa = 11.84LEE371 pKa = 4.43ANNLEE376 pKa = 4.2ATAGSGGAIFYY387 pKa = 10.26VPNGDD392 pKa = 3.51VTIGGVTDD400 pKa = 3.91EE401 pKa = 4.46LTGMSITDD409 pKa = 3.53GGDD412 pKa = 3.27FSLEE416 pKa = 4.17SVGSITLIEE425 pKa = 5.35DD426 pKa = 3.98IYY428 pKa = 11.14TSDD431 pKa = 5.37DD432 pKa = 3.52IQSGDD437 pKa = 3.03ITLTSNAGAIDD448 pKa = 3.78TTGGGISAYY457 pKa = 10.17SEE459 pKa = 4.17YY460 pKa = 11.29GNGGSVSLTAQGDD473 pKa = 3.49IHH475 pKa = 6.45TSNISSYY482 pKa = 10.99SPEE485 pKa = 3.75NGGNVSLEE493 pKa = 4.3STGGAIDD500 pKa = 3.61TTGGEE505 pKa = 3.84ISANSGYY512 pKa = 11.24GNGGSVSLTAQGDD525 pKa = 3.49IHH527 pKa = 6.48TSNISSDD534 pKa = 2.77SWFWTGGNVSLEE546 pKa = 4.31STNGAIDD553 pKa = 3.54TTGGEE558 pKa = 3.75ISANSVSGNAGDD570 pKa = 4.49VSLTAQGDD578 pKa = 3.47IHH580 pKa = 6.36TSNIRR585 pKa = 11.84SYY587 pKa = 11.09SSEE590 pKa = 3.91GNGGNVSLEE599 pKa = 4.27STGGEE604 pKa = 3.69IDD606 pKa = 3.55TTGGGILAGSGSSGGDD622 pKa = 2.99VSLIAQGDD630 pKa = 3.55IHH632 pKa = 6.45TSNIRR637 pKa = 11.84SFSWDD642 pKa = 3.06GTGGNVFLEE651 pKa = 4.52STGGAIDD658 pKa = 3.61TTGGEE663 pKa = 3.94ISPYY667 pKa = 9.72SAYY670 pKa = 10.69GNAGDD675 pKa = 4.44VSLIAQGDD683 pKa = 3.58IHH685 pKa = 6.27TSRR688 pKa = 11.84IGSYY692 pKa = 9.99SGEE695 pKa = 4.05GTGGNVFLEE704 pKa = 4.52STDD707 pKa = 3.63GAIDD711 pKa = 3.55TTGGEE716 pKa = 3.75ISANSVSGNGGSVSLTAQGDD736 pKa = 3.47IHH738 pKa = 6.36TSNIRR743 pKa = 11.84SYY745 pKa = 11.09SSEE748 pKa = 3.91GNGGNVSLEE757 pKa = 4.29STGGAINTSAGQLDD771 pKa = 3.93SHH773 pKa = 5.94SQEE776 pKa = 3.88GSAGEE781 pKa = 4.07IKK783 pKa = 9.92IDD785 pKa = 3.46ASGDD789 pKa = 3.23ILTGFVSSYY798 pKa = 9.68NQSSSADD805 pKa = 3.75SYY807 pKa = 12.0SGDD810 pKa = 2.67IWITSHH816 pKa = 6.5NGSINTTIGNLSGEE830 pKa = 4.33AQISPNDD837 pKa = 3.78DD838 pKa = 3.27VSSVEE843 pKa = 3.98VASIFNNAQANLASYY858 pKa = 9.68AHH860 pKa = 6.91SGTGGNVILKK870 pKa = 10.4ANGDD874 pKa = 3.34ITTSHH879 pKa = 7.1ISTFGSQSSGNVNIISNNGAINTGVIFSFSQLNNAGDD916 pKa = 3.75VTLNAAGNISTSHH929 pKa = 5.68ISAWGNQQGGNITIDD944 pKa = 3.22AGNIAGQINNNTPCDD959 pKa = 3.84CVNLLGTEE967 pKa = 4.08PTPSFNIGSATIQSFSQVGHH987 pKa = 7.03AGNVTITTSGDD998 pKa = 3.24TNLGGDD1004 pKa = 4.26EE1005 pKa = 4.04NRR1007 pKa = 11.84HH1008 pKa = 6.14AIRR1011 pKa = 11.84SAGYY1015 pKa = 8.06TEE1017 pKa = 5.08GGNISIFSLGG1027 pKa = 3.29
Molecular weight: 102.96 kDa
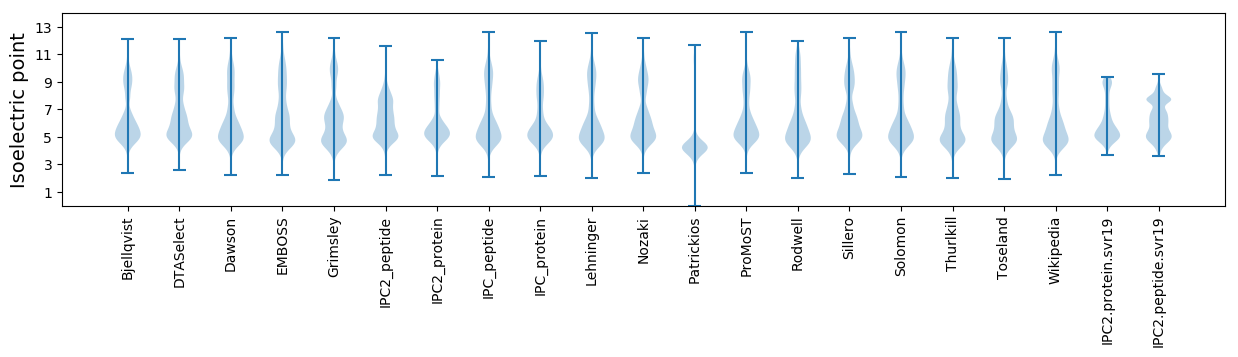
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M1JMQ7|A0A0M1JMQ7_9CYAN AAA family ATPase OS=Planktothricoides sp. SR001 OX=1705388 GN=AM228_23335 PE=4 SV=1
MM1 pKa = 7.59GEE3 pKa = 4.18NVNLNTGMRR12 pKa = 11.84GPRR15 pKa = 11.84YY16 pKa = 9.18LWAKK20 pKa = 9.88ILINIVGAKK29 pKa = 9.95HH30 pKa = 6.55SGRR33 pKa = 11.84CFPGEE38 pKa = 4.11NVNLITGMLRR48 pKa = 11.84PWRR51 pKa = 11.84YY52 pKa = 9.08LWAKK56 pKa = 8.97MLINIVGAKK65 pKa = 9.95HH66 pKa = 6.55SGRR69 pKa = 11.84CFPGEE74 pKa = 3.89NVNLNTGMRR83 pKa = 11.84GPRR86 pKa = 11.84YY87 pKa = 8.94LWAIAMISSRR97 pKa = 11.84CQGEE101 pKa = 3.98AFPP104 pKa = 5.76
MM1 pKa = 7.59GEE3 pKa = 4.18NVNLNTGMRR12 pKa = 11.84GPRR15 pKa = 11.84YY16 pKa = 9.18LWAKK20 pKa = 9.88ILINIVGAKK29 pKa = 9.95HH30 pKa = 6.55SGRR33 pKa = 11.84CFPGEE38 pKa = 4.11NVNLITGMLRR48 pKa = 11.84PWRR51 pKa = 11.84YY52 pKa = 9.08LWAKK56 pKa = 8.97MLINIVGAKK65 pKa = 9.95HH66 pKa = 6.55SGRR69 pKa = 11.84CFPGEE74 pKa = 3.89NVNLNTGMRR83 pKa = 11.84GPRR86 pKa = 11.84YY87 pKa = 8.94LWAIAMISSRR97 pKa = 11.84CQGEE101 pKa = 3.98AFPP104 pKa = 5.76
Molecular weight: 11.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1533545 |
34 |
2694 |
304.2 |
33.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.069 ± 0.036 | 1.008 ± 0.013 |
5.046 ± 0.03 | 6.46 ± 0.032 |
3.953 ± 0.022 | 6.727 ± 0.04 |
1.807 ± 0.019 | 6.925 ± 0.034 |
4.884 ± 0.028 | 10.877 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.989 ± 0.015 | 4.466 ± 0.034 |
4.909 ± 0.028 | 5.43 ± 0.033 |
5.072 ± 0.029 | 6.269 ± 0.027 |
5.41 ± 0.032 | 6.164 ± 0.032 |
1.465 ± 0.016 | 3.069 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
