
Microbacterium sp. SLBN-1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium; unclassified Microbacterium
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
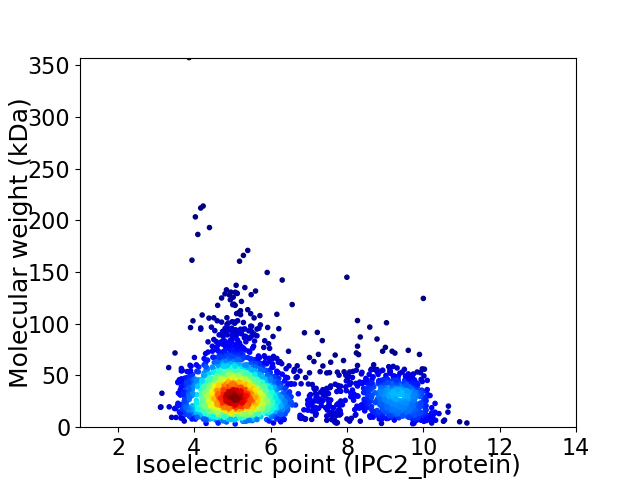
Virtual 2D-PAGE plot for 2917 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A560MNA6|A0A560MNA6_9MICO MFS transporter OS=Microbacterium sp. SLBN-1 OX=2768438 GN=FBY16_1282 PE=4 SV=1
MM1 pKa = 7.59ARR3 pKa = 11.84NTHH6 pKa = 4.75HH7 pKa = 6.54TARR10 pKa = 11.84FGFPGPALRR19 pKa = 11.84RR20 pKa = 11.84AAAGLAVVTVGALALSACTTSTEE43 pKa = 4.67DD44 pKa = 3.37PGTTEE49 pKa = 4.84GGTLKK54 pKa = 10.42LWHH57 pKa = 6.48YY58 pKa = 9.86EE59 pKa = 4.19GADD62 pKa = 3.47SAMGKK67 pKa = 9.56AWAEE71 pKa = 4.07AISVFEE77 pKa = 4.52EE78 pKa = 4.02EE79 pKa = 3.8TGATVEE85 pKa = 4.18FEE87 pKa = 4.46EE88 pKa = 5.81KK89 pKa = 10.51SFEE92 pKa = 4.27QIQSTASQVLDD103 pKa = 3.35TDD105 pKa = 4.22AAPDD109 pKa = 3.44LMEE112 pKa = 4.6FNKK115 pKa = 10.57GNATAGFLASTGLISDD131 pKa = 3.71ISDD134 pKa = 3.35AVDD137 pKa = 3.37EE138 pKa = 4.73YY139 pKa = 11.84GWADD143 pKa = 3.46KK144 pKa = 10.46LAPSLQTTAKK154 pKa = 8.49YY155 pKa = 9.42TDD157 pKa = 4.02DD158 pKa = 3.59GVMGGDD164 pKa = 3.07TWYY167 pKa = 10.81GIPNYY172 pKa = 10.79GEE174 pKa = 4.11FVGVYY179 pKa = 9.96YY180 pKa = 10.98NLDD183 pKa = 3.38AFADD187 pKa = 3.73AGLEE191 pKa = 4.16VPTTYY196 pKa = 11.19DD197 pKa = 2.87EE198 pKa = 4.8FVAVLDD204 pKa = 3.85AFVAKK209 pKa = 10.61GITPLAEE216 pKa = 4.56AGAEE220 pKa = 4.13YY221 pKa = 10.11PLGQLWYY228 pKa = 10.59QLALSQGDD236 pKa = 3.54RR237 pKa = 11.84QFVNDD242 pKa = 3.53YY243 pKa = 10.58QLYY246 pKa = 8.78EE247 pKa = 4.77NPVDD251 pKa = 3.49WQGPEE256 pKa = 3.33ITYY259 pKa = 10.69ASQTLADD266 pKa = 4.15YY267 pKa = 11.04VEE269 pKa = 4.43KK270 pKa = 10.81GYY272 pKa = 10.3IASDD276 pKa = 3.62VSSVKK281 pKa = 10.95AEE283 pKa = 3.91DD284 pKa = 3.76AGVSFINGTAPIFVSGSWWYY304 pKa = 10.97GRR306 pKa = 11.84FVSEE310 pKa = 4.2ATGFDD315 pKa = 3.22WTMTAFPEE323 pKa = 4.33ATLSLGSSGNLWVVPEE339 pKa = 3.88NAANKK344 pKa = 8.56EE345 pKa = 3.81LAYY348 pKa = 10.18EE349 pKa = 4.84FIDD352 pKa = 2.86ITMRR356 pKa = 11.84PEE358 pKa = 3.16IQAIIGNNGGVPVAADD374 pKa = 3.41PADD377 pKa = 3.45ITDD380 pKa = 3.84EE381 pKa = 4.25KK382 pKa = 10.65SQEE385 pKa = 4.36LITVFNGILEE395 pKa = 4.4ADD397 pKa = 3.93GLSFYY402 pKa = 10.21PDD404 pKa = 2.74WPAPGFYY411 pKa = 10.52DD412 pKa = 4.99VIVQQLQGLVTGSQDD427 pKa = 3.5VATTNTNLGAQYY439 pKa = 11.15DD440 pKa = 4.03EE441 pKa = 4.46GTADD445 pKa = 3.63FRR447 pKa = 5.5
MM1 pKa = 7.59ARR3 pKa = 11.84NTHH6 pKa = 4.75HH7 pKa = 6.54TARR10 pKa = 11.84FGFPGPALRR19 pKa = 11.84RR20 pKa = 11.84AAAGLAVVTVGALALSACTTSTEE43 pKa = 4.67DD44 pKa = 3.37PGTTEE49 pKa = 4.84GGTLKK54 pKa = 10.42LWHH57 pKa = 6.48YY58 pKa = 9.86EE59 pKa = 4.19GADD62 pKa = 3.47SAMGKK67 pKa = 9.56AWAEE71 pKa = 4.07AISVFEE77 pKa = 4.52EE78 pKa = 4.02EE79 pKa = 3.8TGATVEE85 pKa = 4.18FEE87 pKa = 4.46EE88 pKa = 5.81KK89 pKa = 10.51SFEE92 pKa = 4.27QIQSTASQVLDD103 pKa = 3.35TDD105 pKa = 4.22AAPDD109 pKa = 3.44LMEE112 pKa = 4.6FNKK115 pKa = 10.57GNATAGFLASTGLISDD131 pKa = 3.71ISDD134 pKa = 3.35AVDD137 pKa = 3.37EE138 pKa = 4.73YY139 pKa = 11.84GWADD143 pKa = 3.46KK144 pKa = 10.46LAPSLQTTAKK154 pKa = 8.49YY155 pKa = 9.42TDD157 pKa = 4.02DD158 pKa = 3.59GVMGGDD164 pKa = 3.07TWYY167 pKa = 10.81GIPNYY172 pKa = 10.79GEE174 pKa = 4.11FVGVYY179 pKa = 9.96YY180 pKa = 10.98NLDD183 pKa = 3.38AFADD187 pKa = 3.73AGLEE191 pKa = 4.16VPTTYY196 pKa = 11.19DD197 pKa = 2.87EE198 pKa = 4.8FVAVLDD204 pKa = 3.85AFVAKK209 pKa = 10.61GITPLAEE216 pKa = 4.56AGAEE220 pKa = 4.13YY221 pKa = 10.11PLGQLWYY228 pKa = 10.59QLALSQGDD236 pKa = 3.54RR237 pKa = 11.84QFVNDD242 pKa = 3.53YY243 pKa = 10.58QLYY246 pKa = 8.78EE247 pKa = 4.77NPVDD251 pKa = 3.49WQGPEE256 pKa = 3.33ITYY259 pKa = 10.69ASQTLADD266 pKa = 4.15YY267 pKa = 11.04VEE269 pKa = 4.43KK270 pKa = 10.81GYY272 pKa = 10.3IASDD276 pKa = 3.62VSSVKK281 pKa = 10.95AEE283 pKa = 3.91DD284 pKa = 3.76AGVSFINGTAPIFVSGSWWYY304 pKa = 10.97GRR306 pKa = 11.84FVSEE310 pKa = 4.2ATGFDD315 pKa = 3.22WTMTAFPEE323 pKa = 4.33ATLSLGSSGNLWVVPEE339 pKa = 3.88NAANKK344 pKa = 8.56EE345 pKa = 3.81LAYY348 pKa = 10.18EE349 pKa = 4.84FIDD352 pKa = 2.86ITMRR356 pKa = 11.84PEE358 pKa = 3.16IQAIIGNNGGVPVAADD374 pKa = 3.41PADD377 pKa = 3.45ITDD380 pKa = 3.84EE381 pKa = 4.25KK382 pKa = 10.65SQEE385 pKa = 4.36LITVFNGILEE395 pKa = 4.4ADD397 pKa = 3.93GLSFYY402 pKa = 10.21PDD404 pKa = 2.74WPAPGFYY411 pKa = 10.52DD412 pKa = 4.99VIVQQLQGLVTGSQDD427 pKa = 3.5VATTNTNLGAQYY439 pKa = 11.15DD440 pKa = 4.03EE441 pKa = 4.46GTADD445 pKa = 3.63FRR447 pKa = 5.5
Molecular weight: 47.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A560MKL4|A0A560MKL4_9MICO DUF1508 domain-containing protein OS=Microbacterium sp. SLBN-1 OX=2768438 GN=FBY16_0316 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
977528 |
29 |
3537 |
335.1 |
35.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.898 ± 0.068 | 0.457 ± 0.011 |
6.535 ± 0.042 | 5.462 ± 0.039 |
3.116 ± 0.029 | 8.832 ± 0.043 |
1.978 ± 0.024 | 4.321 ± 0.033 |
1.796 ± 0.029 | 10.057 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.677 ± 0.02 | 1.792 ± 0.027 |
5.403 ± 0.031 | 2.622 ± 0.02 |
7.487 ± 0.057 | 5.573 ± 0.035 |
6.259 ± 0.054 | 9.201 ± 0.052 |
1.565 ± 0.025 | 1.971 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
