
Chaetomium globosum (strain ATCC 6205 / CBS 148.51 / DSM 1962 / NBRC 6347 / NRRL 1970) (Soil fungus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Sordariomycetidae; Sordariales; Chaetomiaceae; Chaetomium; Chaetomium globosum
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
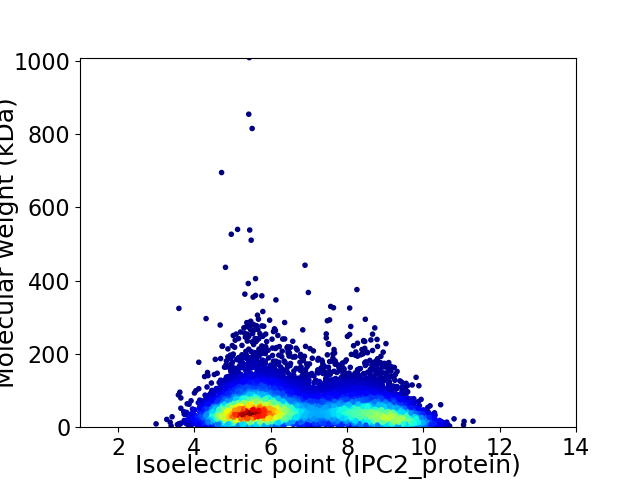
Virtual 2D-PAGE plot for 11042 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q2HEQ2|Q2HEQ2_CHAGB Peptidyl-prolyl cis-trans isomerase OS=Chaetomium globosum (strain ATCC 6205 / CBS 148.51 / DSM 1962 / NBRC 6347 / NRRL 1970) OX=306901 GN=CHGG_01302 PE=3 SV=1
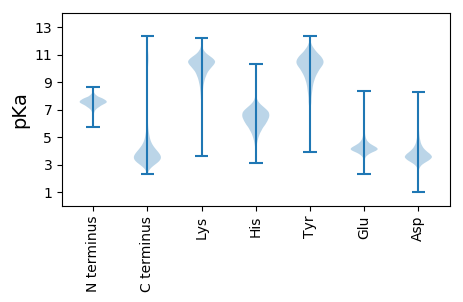
MM1 pKa = 7.14HH2 pKa = 7.3QYY4 pKa = 11.22ACLVSLLPLVAAHH17 pKa = 7.03GFLSSPPARR26 pKa = 11.84RR27 pKa = 11.84PGAAYY32 pKa = 9.83RR33 pKa = 11.84ATCGEE38 pKa = 3.96QPFYY42 pKa = 10.71QQSSDD47 pKa = 3.14INGNVQGIQQVVGSDD62 pKa = 3.05ATEE65 pKa = 4.24DD66 pKa = 3.76CNLWLCKK73 pKa = 10.52GFLLEE78 pKa = 5.45DD79 pKa = 4.14NPDD82 pKa = 3.47QVQSYY87 pKa = 11.22SLGQTIDD94 pKa = 2.98ITVNIAAPHH103 pKa = 5.52TGYY106 pKa = 11.49ANVSVVKK113 pKa = 9.97TSTNTMIGNPLIEE126 pKa = 4.37FQNYY130 pKa = 9.4ASNSGPGPPTTQPSVSPFRR149 pKa = 11.84KK150 pKa = 9.37AWAATAQPLATACSSGSGMPRR171 pKa = 11.84TLIRR175 pKa = 11.84PTRR178 pKa = 11.84PAPTSSWPLTGMGIALSSTSTVPVTTASAPATATSVDD215 pKa = 3.96PAPATTTTAVPGTPDD230 pKa = 4.07DD231 pKa = 4.25EE232 pKa = 5.45CPADD236 pKa = 4.09DD237 pKa = 5.67GGDD240 pKa = 3.56EE241 pKa = 5.57GEE243 pKa = 5.8DD244 pKa = 3.47EE245 pKa = 5.54DD246 pKa = 6.19DD247 pKa = 3.83EE248 pKa = 4.51QGEE251 pKa = 4.37SGEE254 pKa = 4.28GDD256 pKa = 3.54EE257 pKa = 4.81EE258 pKa = 5.27CPVDD262 pKa = 4.15GGEE265 pKa = 4.64EE266 pKa = 4.14PEE268 pKa = 4.62EE269 pKa = 5.75DD270 pKa = 5.26DD271 pKa = 6.77DD272 pKa = 5.92STDD275 pKa = 4.43DD276 pKa = 5.89DD277 pKa = 4.41NAEE280 pKa = 4.36CPADD284 pKa = 4.58DD285 pKa = 5.12GDD287 pKa = 3.95NAAATTSLGAKK298 pKa = 9.68DD299 pKa = 4.04VSTSAAATPTGSHH312 pKa = 6.43TSTVTQIVTSTVTTEE327 pKa = 3.9RR328 pKa = 11.84LVTVTAAATACNN340 pKa = 3.79
MM1 pKa = 7.14HH2 pKa = 7.3QYY4 pKa = 11.22ACLVSLLPLVAAHH17 pKa = 7.03GFLSSPPARR26 pKa = 11.84RR27 pKa = 11.84PGAAYY32 pKa = 9.83RR33 pKa = 11.84ATCGEE38 pKa = 3.96QPFYY42 pKa = 10.71QQSSDD47 pKa = 3.14INGNVQGIQQVVGSDD62 pKa = 3.05ATEE65 pKa = 4.24DD66 pKa = 3.76CNLWLCKK73 pKa = 10.52GFLLEE78 pKa = 5.45DD79 pKa = 4.14NPDD82 pKa = 3.47QVQSYY87 pKa = 11.22SLGQTIDD94 pKa = 2.98ITVNIAAPHH103 pKa = 5.52TGYY106 pKa = 11.49ANVSVVKK113 pKa = 9.97TSTNTMIGNPLIEE126 pKa = 4.37FQNYY130 pKa = 9.4ASNSGPGPPTTQPSVSPFRR149 pKa = 11.84KK150 pKa = 9.37AWAATAQPLATACSSGSGMPRR171 pKa = 11.84TLIRR175 pKa = 11.84PTRR178 pKa = 11.84PAPTSSWPLTGMGIALSSTSTVPVTTASAPATATSVDD215 pKa = 3.96PAPATTTTAVPGTPDD230 pKa = 4.07DD231 pKa = 4.25EE232 pKa = 5.45CPADD236 pKa = 4.09DD237 pKa = 5.67GGDD240 pKa = 3.56EE241 pKa = 5.57GEE243 pKa = 5.8DD244 pKa = 3.47EE245 pKa = 5.54DD246 pKa = 6.19DD247 pKa = 3.83EE248 pKa = 4.51QGEE251 pKa = 4.37SGEE254 pKa = 4.28GDD256 pKa = 3.54EE257 pKa = 4.81EE258 pKa = 5.27CPVDD262 pKa = 4.15GGEE265 pKa = 4.64EE266 pKa = 4.14PEE268 pKa = 4.62EE269 pKa = 5.75DD270 pKa = 5.26DD271 pKa = 6.77DD272 pKa = 5.92STDD275 pKa = 4.43DD276 pKa = 5.89DD277 pKa = 4.41NAEE280 pKa = 4.36CPADD284 pKa = 4.58DD285 pKa = 5.12GDD287 pKa = 3.95NAAATTSLGAKK298 pKa = 9.68DD299 pKa = 4.04VSTSAAATPTGSHH312 pKa = 6.43TSTVTQIVTSTVTTEE327 pKa = 3.9RR328 pKa = 11.84LVTVTAAATACNN340 pKa = 3.79
Molecular weight: 34.82 kDa
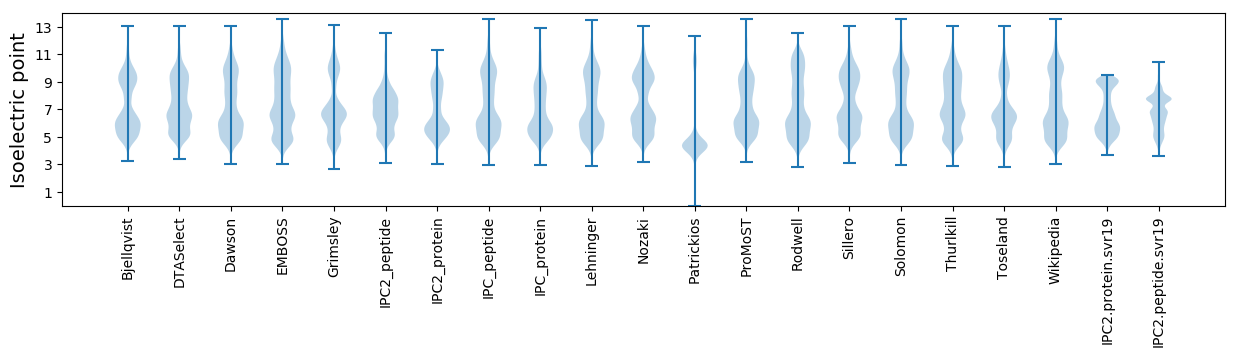
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q2GSD5|Q2GSD5_CHAGB J domain-containing protein OS=Chaetomium globosum (strain ATCC 6205 / CBS 148.51 / DSM 1962 / NBRC 6347 / NRRL 1970) OX=306901 GN=CHGG_09119 PE=4 SV=1
MM1 pKa = 7.87PPPSPTTNQIRR12 pKa = 11.84PHH14 pKa = 5.97SAAPTAPQRR23 pKa = 11.84PPNATTATTGNSHH36 pKa = 7.14PRR38 pKa = 11.84PTISTATHH46 pKa = 4.5QTPSRR51 pKa = 11.84SGTPTTPQPPPNAIIATTGNNNNHH75 pKa = 6.23ARR77 pKa = 11.84PTTGTGNASGAHH89 pKa = 5.02TAVSRR94 pKa = 11.84QGTATPGRR102 pKa = 11.84NGTGAPSPPRR112 pKa = 11.84TNPAPGPNPPPGPGPNQTANLATRR136 pKa = 11.84SANGNGRR143 pKa = 11.84GGGAPGNGNGSATQQQAGGRR163 pKa = 11.84GARR166 pKa = 11.84RR167 pKa = 3.24
MM1 pKa = 7.87PPPSPTTNQIRR12 pKa = 11.84PHH14 pKa = 5.97SAAPTAPQRR23 pKa = 11.84PPNATTATTGNSHH36 pKa = 7.14PRR38 pKa = 11.84PTISTATHH46 pKa = 4.5QTPSRR51 pKa = 11.84SGTPTTPQPPPNAIIATTGNNNNHH75 pKa = 6.23ARR77 pKa = 11.84PTTGTGNASGAHH89 pKa = 5.02TAVSRR94 pKa = 11.84QGTATPGRR102 pKa = 11.84NGTGAPSPPRR112 pKa = 11.84TNPAPGPNPPPGPGPNQTANLATRR136 pKa = 11.84SANGNGRR143 pKa = 11.84GGGAPGNGNGSATQQQAGGRR163 pKa = 11.84GARR166 pKa = 11.84RR167 pKa = 3.24
Molecular weight: 16.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5473786 |
20 |
9223 |
495.7 |
54.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.482 ± 0.021 | 1.24 ± 0.009 |
5.623 ± 0.016 | 6.092 ± 0.026 |
3.413 ± 0.013 | 7.624 ± 0.025 |
2.439 ± 0.011 | 4.183 ± 0.014 |
4.44 ± 0.02 | 8.689 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.072 ± 0.009 | 3.365 ± 0.011 |
6.715 ± 0.028 | 4.013 ± 0.018 |
6.781 ± 0.021 | 7.572 ± 0.024 |
6.07 ± 0.017 | 6.205 ± 0.019 |
1.495 ± 0.009 | 2.474 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
