
Flavobacterium croceum DSM 17960
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium; Flavobacterium croceum
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
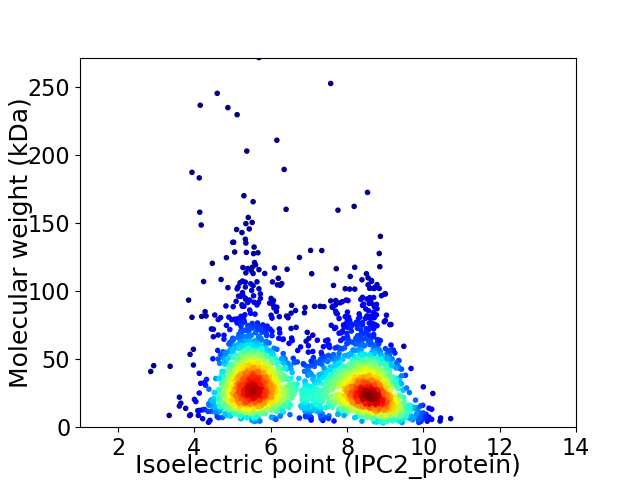
Virtual 2D-PAGE plot for 2701 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
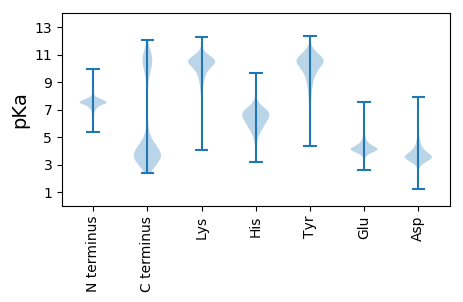
Protein with the lowest isoelectric point:
>tr|A0A2S4N9F2|A0A2S4N9F2_9FLAO Arabinose-5-phosphate isomerase OS=Flavobacterium croceum DSM 17960 OX=1121886 GN=Q361_10424 PE=3 SV=1
MM1 pKa = 7.57KK2 pKa = 10.43KK3 pKa = 10.44LYY5 pKa = 10.47LQILLFFIGLATTVAQVPNYY25 pKa = 9.31NVVSIPYY32 pKa = 9.1QNVVLSPTMASTTISFNLDD51 pKa = 3.14DD52 pKa = 4.76TYY54 pKa = 11.39SGPIALPFNINFYY67 pKa = 11.05GVTNNFMNVSSNGYY81 pKa = 7.95ITFNSNAAGSNSPWSFNTTIPNAGFPVKK109 pKa = 10.41NAFLGCYY116 pKa = 9.53HH117 pKa = 7.79DD118 pKa = 4.3INNAIVNTTDD128 pKa = 3.38PQSVVYY134 pKa = 10.12SIQGYY139 pKa = 9.47APFRR143 pKa = 11.84KK144 pKa = 10.18VIVLYY149 pKa = 10.99SNNPQFSCNNSAISTFAMIMYY170 pKa = 10.14EE171 pKa = 4.17DD172 pKa = 4.35GEE174 pKa = 4.39KK175 pKa = 10.44FDD177 pKa = 3.92VQIKK181 pKa = 10.56RR182 pKa = 11.84KK183 pKa = 9.65DD184 pKa = 3.41LCATWNGGRR193 pKa = 11.84AVVGLIDD200 pKa = 3.71PTGASAIAAPGRR212 pKa = 11.84NTSAWTANYY221 pKa = 7.53EE222 pKa = 3.53AWRR225 pKa = 11.84FYY227 pKa = 10.6NTFGANHH234 pKa = 5.8YY235 pKa = 9.9KK236 pKa = 10.12FWKK239 pKa = 10.38CEE241 pKa = 3.7TDD243 pKa = 3.48GDD245 pKa = 4.1GLEE248 pKa = 4.35PFNLAVVKK256 pKa = 10.06TDD258 pKa = 3.61LNEE261 pKa = 4.11PSMTFYY267 pKa = 11.04SGIINGQVDD276 pKa = 4.42VNTALPEE283 pKa = 4.63LYY285 pKa = 10.65QNTTAFNQKK294 pKa = 9.62IYY296 pKa = 10.48GLKK299 pKa = 8.58STGEE303 pKa = 3.92IVTIDD308 pKa = 3.94LKK310 pKa = 11.47VFSCDD315 pKa = 3.53NDD317 pKa = 3.37YY318 pKa = 11.79DD319 pKa = 4.54LDD321 pKa = 3.99NVTTSLEE328 pKa = 4.02DD329 pKa = 3.86LNNDD333 pKa = 3.34TNLANDD339 pKa = 4.01DD340 pKa = 3.77TDD342 pKa = 5.62ADD344 pKa = 4.0GTPNFIDD351 pKa = 4.09NDD353 pKa = 4.04DD354 pKa = 4.81DD355 pKa = 5.72GDD357 pKa = 4.9LILTSIEE364 pKa = 4.06YY365 pKa = 10.55VFNSNKK371 pKa = 8.06NTNATSTTNSVLDD384 pKa = 3.98TDD386 pKa = 4.19NDD388 pKa = 4.87GIPNYY393 pKa = 10.35LDD395 pKa = 4.14NDD397 pKa = 4.19DD398 pKa = 5.75DD399 pKa = 5.99GDD401 pKa = 4.24GVLTMNEE408 pKa = 4.31DD409 pKa = 3.79YY410 pKa = 11.21NGNRR414 pKa = 11.84NPADD418 pKa = 4.5DD419 pKa = 4.24DD420 pKa = 4.38TNGNSIPDD428 pKa = 3.7YY429 pKa = 11.02LEE431 pKa = 3.84TSVALGTAQNILDD444 pKa = 4.26SYY446 pKa = 9.05FTVYY450 pKa = 10.42PNPANDD456 pKa = 3.42VLFIKK461 pKa = 10.72NNNFNNADD469 pKa = 3.33IKK471 pKa = 11.09VKK473 pKa = 10.14IVSVTGAVVKK483 pKa = 10.52EE484 pKa = 3.92MSTQNEE490 pKa = 3.96ITPISVSALQSGMYY504 pKa = 9.2IVEE507 pKa = 5.08AISSEE512 pKa = 3.74GTSYY516 pKa = 11.29HH517 pKa = 6.81KK518 pKa = 10.45FIKK521 pKa = 10.27KK522 pKa = 9.85
MM1 pKa = 7.57KK2 pKa = 10.43KK3 pKa = 10.44LYY5 pKa = 10.47LQILLFFIGLATTVAQVPNYY25 pKa = 9.31NVVSIPYY32 pKa = 9.1QNVVLSPTMASTTISFNLDD51 pKa = 3.14DD52 pKa = 4.76TYY54 pKa = 11.39SGPIALPFNINFYY67 pKa = 11.05GVTNNFMNVSSNGYY81 pKa = 7.95ITFNSNAAGSNSPWSFNTTIPNAGFPVKK109 pKa = 10.41NAFLGCYY116 pKa = 9.53HH117 pKa = 7.79DD118 pKa = 4.3INNAIVNTTDD128 pKa = 3.38PQSVVYY134 pKa = 10.12SIQGYY139 pKa = 9.47APFRR143 pKa = 11.84KK144 pKa = 10.18VIVLYY149 pKa = 10.99SNNPQFSCNNSAISTFAMIMYY170 pKa = 10.14EE171 pKa = 4.17DD172 pKa = 4.35GEE174 pKa = 4.39KK175 pKa = 10.44FDD177 pKa = 3.92VQIKK181 pKa = 10.56RR182 pKa = 11.84KK183 pKa = 9.65DD184 pKa = 3.41LCATWNGGRR193 pKa = 11.84AVVGLIDD200 pKa = 3.71PTGASAIAAPGRR212 pKa = 11.84NTSAWTANYY221 pKa = 7.53EE222 pKa = 3.53AWRR225 pKa = 11.84FYY227 pKa = 10.6NTFGANHH234 pKa = 5.8YY235 pKa = 9.9KK236 pKa = 10.12FWKK239 pKa = 10.38CEE241 pKa = 3.7TDD243 pKa = 3.48GDD245 pKa = 4.1GLEE248 pKa = 4.35PFNLAVVKK256 pKa = 10.06TDD258 pKa = 3.61LNEE261 pKa = 4.11PSMTFYY267 pKa = 11.04SGIINGQVDD276 pKa = 4.42VNTALPEE283 pKa = 4.63LYY285 pKa = 10.65QNTTAFNQKK294 pKa = 9.62IYY296 pKa = 10.48GLKK299 pKa = 8.58STGEE303 pKa = 3.92IVTIDD308 pKa = 3.94LKK310 pKa = 11.47VFSCDD315 pKa = 3.53NDD317 pKa = 3.37YY318 pKa = 11.79DD319 pKa = 4.54LDD321 pKa = 3.99NVTTSLEE328 pKa = 4.02DD329 pKa = 3.86LNNDD333 pKa = 3.34TNLANDD339 pKa = 4.01DD340 pKa = 3.77TDD342 pKa = 5.62ADD344 pKa = 4.0GTPNFIDD351 pKa = 4.09NDD353 pKa = 4.04DD354 pKa = 4.81DD355 pKa = 5.72GDD357 pKa = 4.9LILTSIEE364 pKa = 4.06YY365 pKa = 10.55VFNSNKK371 pKa = 8.06NTNATSTTNSVLDD384 pKa = 3.98TDD386 pKa = 4.19NDD388 pKa = 4.87GIPNYY393 pKa = 10.35LDD395 pKa = 4.14NDD397 pKa = 4.19DD398 pKa = 5.75DD399 pKa = 5.99GDD401 pKa = 4.24GVLTMNEE408 pKa = 4.31DD409 pKa = 3.79YY410 pKa = 11.21NGNRR414 pKa = 11.84NPADD418 pKa = 4.5DD419 pKa = 4.24DD420 pKa = 4.38TNGNSIPDD428 pKa = 3.7YY429 pKa = 11.02LEE431 pKa = 3.84TSVALGTAQNILDD444 pKa = 4.26SYY446 pKa = 9.05FTVYY450 pKa = 10.42PNPANDD456 pKa = 3.42VLFIKK461 pKa = 10.72NNNFNNADD469 pKa = 3.33IKK471 pKa = 11.09VKK473 pKa = 10.14IVSVTGAVVKK483 pKa = 10.52EE484 pKa = 3.92MSTQNEE490 pKa = 3.96ITPISVSALQSGMYY504 pKa = 9.2IVEE507 pKa = 5.08AISSEE512 pKa = 3.74GTSYY516 pKa = 11.29HH517 pKa = 6.81KK518 pKa = 10.45FIKK521 pKa = 10.27KK522 pKa = 9.85
Molecular weight: 57.26 kDa
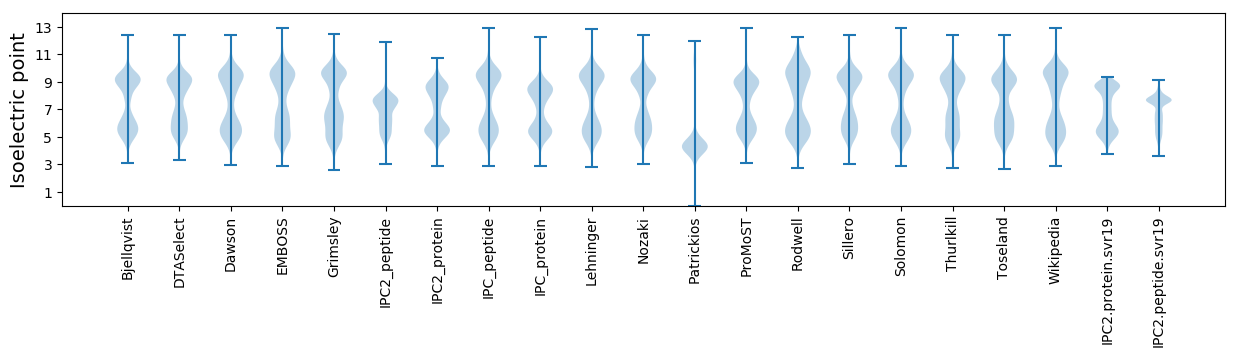
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S4N809|A0A2S4N809_9FLAO Phosphoribosyl pyrophosphate synthase OS=Flavobacterium croceum DSM 17960 OX=1121886 GN=Q361_10721 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.53HH17 pKa = 4.24GFMEE21 pKa = 5.09RR22 pKa = 11.84MATANGRR29 pKa = 11.84KK30 pKa = 8.93VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.21GRR40 pKa = 11.84HH41 pKa = 5.23KK42 pKa = 10.67LSVSSEE48 pKa = 3.91PRR50 pKa = 11.84HH51 pKa = 5.92KK52 pKa = 10.61KK53 pKa = 9.84
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.53HH17 pKa = 4.24GFMEE21 pKa = 5.09RR22 pKa = 11.84MATANGRR29 pKa = 11.84KK30 pKa = 8.93VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.21GRR40 pKa = 11.84HH41 pKa = 5.23KK42 pKa = 10.67LSVSSEE48 pKa = 3.91PRR50 pKa = 11.84HH51 pKa = 5.92KK52 pKa = 10.61KK53 pKa = 9.84
Molecular weight: 6.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
889783 |
28 |
2406 |
329.4 |
37.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.067 ± 0.054 | 0.86 ± 0.015 |
4.966 ± 0.038 | 6.247 ± 0.057 |
5.359 ± 0.038 | 5.822 ± 0.045 |
1.753 ± 0.024 | 8.215 ± 0.048 |
8.383 ± 0.065 | 9.079 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.115 ± 0.027 | 6.841 ± 0.062 |
3.281 ± 0.026 | 3.773 ± 0.031 |
2.859 ± 0.036 | 6.469 ± 0.048 |
6.34 ± 0.076 | 6.151 ± 0.038 |
0.984 ± 0.015 | 4.434 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
