
Bifidobacterium eulemuris
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Bifidobacteriales; Bifidobacteriaceae; Bifidobacterium
Average proteome isoelectric point is 5.83
Get precalculated fractions of proteins
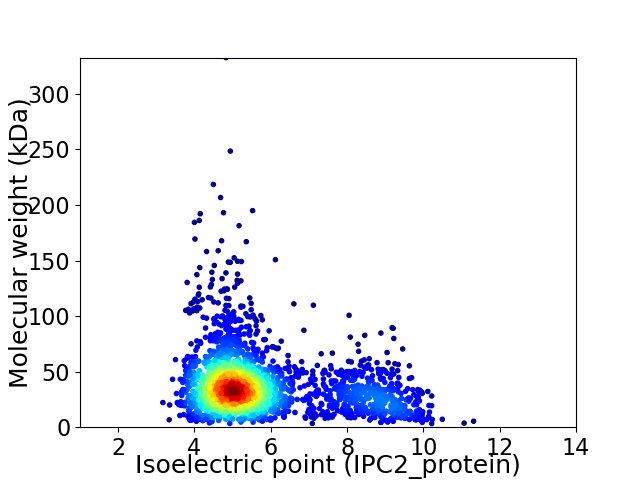
Virtual 2D-PAGE plot for 2326 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A261GDI2|A0A261GDI2_9BIFI Uncharacterized protein OS=Bifidobacterium eulemuris OX=1765219 GN=BEUL_0719 PE=4 SV=1
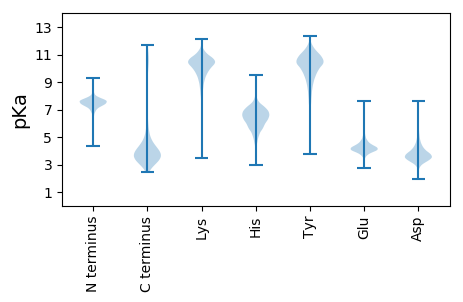
MM1 pKa = 7.51KK2 pKa = 10.12RR3 pKa = 11.84GKK5 pKa = 8.58LTAVVAFTVSAMLALSGCGGSDD27 pKa = 3.16TATDD31 pKa = 3.41AVDD34 pKa = 3.35NGEE37 pKa = 4.65EE38 pKa = 4.03ITLDD42 pKa = 3.7VYY44 pKa = 11.81DD45 pKa = 5.59DD46 pKa = 3.7FANYY50 pKa = 10.25AGEE53 pKa = 4.15QKK55 pKa = 10.7GWFAQVIKK63 pKa = 10.94DD64 pKa = 3.64KK65 pKa = 11.38FNIKK69 pKa = 10.49LNIIAPNVSGGGSTLFDD86 pKa = 3.35TRR88 pKa = 11.84SAAGDD93 pKa = 3.52LGDD96 pKa = 3.65IVLYY100 pKa = 10.25NANNHH105 pKa = 4.97STADD109 pKa = 4.18LIKK112 pKa = 10.65AGLIADD118 pKa = 4.06MTPYY122 pKa = 10.82LDD124 pKa = 3.67DD125 pKa = 4.34AEE127 pKa = 4.7YY128 pKa = 10.4IKK130 pKa = 10.56QYY132 pKa = 11.36SSATDD137 pKa = 3.59SINEE141 pKa = 3.95VAGQEE146 pKa = 4.15SGVWGIPGSVSAQSPEE162 pKa = 3.94TSSEE166 pKa = 3.97GNEE169 pKa = 4.08PTFGPYY175 pKa = 9.36IRR177 pKa = 11.84WDD179 pKa = 3.51YY180 pKa = 9.88YY181 pKa = 11.74AEE183 pKa = 3.98IGYY186 pKa = 9.04PEE188 pKa = 4.75IKK190 pKa = 10.88DD191 pKa = 3.6MDD193 pKa = 4.53DD194 pKa = 3.85LLDD197 pKa = 3.71VLEE200 pKa = 4.33QMQEE204 pKa = 3.68QARR207 pKa = 11.84ADD209 pKa = 3.63TGSTDD214 pKa = 3.27VYY216 pKa = 11.22ALSLFKK222 pKa = 10.93DD223 pKa = 3.2WDD225 pKa = 4.07DD226 pKa = 4.93NMMNNAKK233 pKa = 9.96QPTCFYY239 pKa = 10.68GFDD242 pKa = 3.32EE243 pKa = 4.63MGFVLAKK250 pKa = 10.47ADD252 pKa = 3.53GSDD255 pKa = 3.71YY256 pKa = 11.3EE257 pKa = 4.6DD258 pKa = 3.55VSEE261 pKa = 4.38EE262 pKa = 3.98GGIYY266 pKa = 10.11EE267 pKa = 3.98QTLEE271 pKa = 4.56FFNQAYY277 pKa = 9.53QRR279 pKa = 11.84GLVDD283 pKa = 4.55PEE285 pKa = 5.06SSTQSYY291 pKa = 8.29DD292 pKa = 2.94TMYY295 pKa = 11.42AKK297 pKa = 10.49YY298 pKa = 10.17QEE300 pKa = 4.49GNVLFSFWPWLGQAAYY316 pKa = 7.96NTTEE320 pKa = 3.81NKK322 pKa = 10.28AAGKK326 pKa = 10.63GFMLAPLEE334 pKa = 4.24DD335 pKa = 3.81MQIFSYY341 pKa = 10.71GATPNGGTYY350 pKa = 10.5VIALGAKK357 pKa = 9.46AEE359 pKa = 4.06YY360 pKa = 9.76KK361 pKa = 9.8QRR363 pKa = 11.84CVDD366 pKa = 4.37FINWLYY372 pKa = 10.9SPEE375 pKa = 4.43GIYY378 pKa = 10.72ASGGQTSGSSAGVEE392 pKa = 4.01GLTWEE397 pKa = 4.78LNDD400 pKa = 4.84DD401 pKa = 4.32GEE403 pKa = 4.64PVLTDD408 pKa = 3.27FGYY411 pKa = 9.62QVKK414 pKa = 9.78YY415 pKa = 10.91GSGGTVPDD423 pKa = 3.99EE424 pKa = 4.27YY425 pKa = 11.53GGGNYY430 pKa = 9.16TDD432 pKa = 4.57GVSALNFVTVLASDD446 pKa = 3.97IDD448 pKa = 4.14PEE450 pKa = 4.02TGYY453 pKa = 11.29AYY455 pKa = 10.06DD456 pKa = 3.6ASLWPSVIEE465 pKa = 4.38SQFSTALDD473 pKa = 3.95NDD475 pKa = 3.77WSEE478 pKa = 4.31HH479 pKa = 4.91MGGAKK484 pKa = 8.84TAMEE488 pKa = 4.04YY489 pKa = 11.16LEE491 pKa = 5.54ANDD494 pKa = 5.17KK495 pKa = 11.01ILVAPGASFVKK506 pKa = 10.62PEE508 pKa = 4.86DD509 pKa = 4.0DD510 pKa = 3.85SQISTLRR517 pKa = 11.84GQLKK521 pKa = 6.95TTVVNASWQAVFASSDD537 pKa = 3.79SEE539 pKa = 4.21FQSILDD545 pKa = 3.64GMRR548 pKa = 11.84TDD550 pKa = 5.38LEE552 pKa = 4.28GLGYY556 pKa = 10.58DD557 pKa = 3.4QVLEE561 pKa = 4.03VDD563 pKa = 4.0MKK565 pKa = 10.95NAQDD569 pKa = 3.02QDD571 pKa = 3.49AARR574 pKa = 11.84KK575 pKa = 9.67QIVEE579 pKa = 4.75DD580 pKa = 3.9YY581 pKa = 10.81QSEE584 pKa = 4.26NN585 pKa = 3.14
MM1 pKa = 7.51KK2 pKa = 10.12RR3 pKa = 11.84GKK5 pKa = 8.58LTAVVAFTVSAMLALSGCGGSDD27 pKa = 3.16TATDD31 pKa = 3.41AVDD34 pKa = 3.35NGEE37 pKa = 4.65EE38 pKa = 4.03ITLDD42 pKa = 3.7VYY44 pKa = 11.81DD45 pKa = 5.59DD46 pKa = 3.7FANYY50 pKa = 10.25AGEE53 pKa = 4.15QKK55 pKa = 10.7GWFAQVIKK63 pKa = 10.94DD64 pKa = 3.64KK65 pKa = 11.38FNIKK69 pKa = 10.49LNIIAPNVSGGGSTLFDD86 pKa = 3.35TRR88 pKa = 11.84SAAGDD93 pKa = 3.52LGDD96 pKa = 3.65IVLYY100 pKa = 10.25NANNHH105 pKa = 4.97STADD109 pKa = 4.18LIKK112 pKa = 10.65AGLIADD118 pKa = 4.06MTPYY122 pKa = 10.82LDD124 pKa = 3.67DD125 pKa = 4.34AEE127 pKa = 4.7YY128 pKa = 10.4IKK130 pKa = 10.56QYY132 pKa = 11.36SSATDD137 pKa = 3.59SINEE141 pKa = 3.95VAGQEE146 pKa = 4.15SGVWGIPGSVSAQSPEE162 pKa = 3.94TSSEE166 pKa = 3.97GNEE169 pKa = 4.08PTFGPYY175 pKa = 9.36IRR177 pKa = 11.84WDD179 pKa = 3.51YY180 pKa = 9.88YY181 pKa = 11.74AEE183 pKa = 3.98IGYY186 pKa = 9.04PEE188 pKa = 4.75IKK190 pKa = 10.88DD191 pKa = 3.6MDD193 pKa = 4.53DD194 pKa = 3.85LLDD197 pKa = 3.71VLEE200 pKa = 4.33QMQEE204 pKa = 3.68QARR207 pKa = 11.84ADD209 pKa = 3.63TGSTDD214 pKa = 3.27VYY216 pKa = 11.22ALSLFKK222 pKa = 10.93DD223 pKa = 3.2WDD225 pKa = 4.07DD226 pKa = 4.93NMMNNAKK233 pKa = 9.96QPTCFYY239 pKa = 10.68GFDD242 pKa = 3.32EE243 pKa = 4.63MGFVLAKK250 pKa = 10.47ADD252 pKa = 3.53GSDD255 pKa = 3.71YY256 pKa = 11.3EE257 pKa = 4.6DD258 pKa = 3.55VSEE261 pKa = 4.38EE262 pKa = 3.98GGIYY266 pKa = 10.11EE267 pKa = 3.98QTLEE271 pKa = 4.56FFNQAYY277 pKa = 9.53QRR279 pKa = 11.84GLVDD283 pKa = 4.55PEE285 pKa = 5.06SSTQSYY291 pKa = 8.29DD292 pKa = 2.94TMYY295 pKa = 11.42AKK297 pKa = 10.49YY298 pKa = 10.17QEE300 pKa = 4.49GNVLFSFWPWLGQAAYY316 pKa = 7.96NTTEE320 pKa = 3.81NKK322 pKa = 10.28AAGKK326 pKa = 10.63GFMLAPLEE334 pKa = 4.24DD335 pKa = 3.81MQIFSYY341 pKa = 10.71GATPNGGTYY350 pKa = 10.5VIALGAKK357 pKa = 9.46AEE359 pKa = 4.06YY360 pKa = 9.76KK361 pKa = 9.8QRR363 pKa = 11.84CVDD366 pKa = 4.37FINWLYY372 pKa = 10.9SPEE375 pKa = 4.43GIYY378 pKa = 10.72ASGGQTSGSSAGVEE392 pKa = 4.01GLTWEE397 pKa = 4.78LNDD400 pKa = 4.84DD401 pKa = 4.32GEE403 pKa = 4.64PVLTDD408 pKa = 3.27FGYY411 pKa = 9.62QVKK414 pKa = 9.78YY415 pKa = 10.91GSGGTVPDD423 pKa = 3.99EE424 pKa = 4.27YY425 pKa = 11.53GGGNYY430 pKa = 9.16TDD432 pKa = 4.57GVSALNFVTVLASDD446 pKa = 3.97IDD448 pKa = 4.14PEE450 pKa = 4.02TGYY453 pKa = 11.29AYY455 pKa = 10.06DD456 pKa = 3.6ASLWPSVIEE465 pKa = 4.38SQFSTALDD473 pKa = 3.95NDD475 pKa = 3.77WSEE478 pKa = 4.31HH479 pKa = 4.91MGGAKK484 pKa = 8.84TAMEE488 pKa = 4.04YY489 pKa = 11.16LEE491 pKa = 5.54ANDD494 pKa = 5.17KK495 pKa = 11.01ILVAPGASFVKK506 pKa = 10.62PEE508 pKa = 4.86DD509 pKa = 4.0DD510 pKa = 3.85SQISTLRR517 pKa = 11.84GQLKK521 pKa = 6.95TTVVNASWQAVFASSDD537 pKa = 3.79SEE539 pKa = 4.21FQSILDD545 pKa = 3.64GMRR548 pKa = 11.84TDD550 pKa = 5.38LEE552 pKa = 4.28GLGYY556 pKa = 10.58DD557 pKa = 3.4QVLEE561 pKa = 4.03VDD563 pKa = 4.0MKK565 pKa = 10.95NAQDD569 pKa = 3.02QDD571 pKa = 3.49AARR574 pKa = 11.84KK575 pKa = 9.67QIVEE579 pKa = 4.75DD580 pKa = 3.9YY581 pKa = 10.81QSEE584 pKa = 4.26NN585 pKa = 3.14
Molecular weight: 63.64 kDa
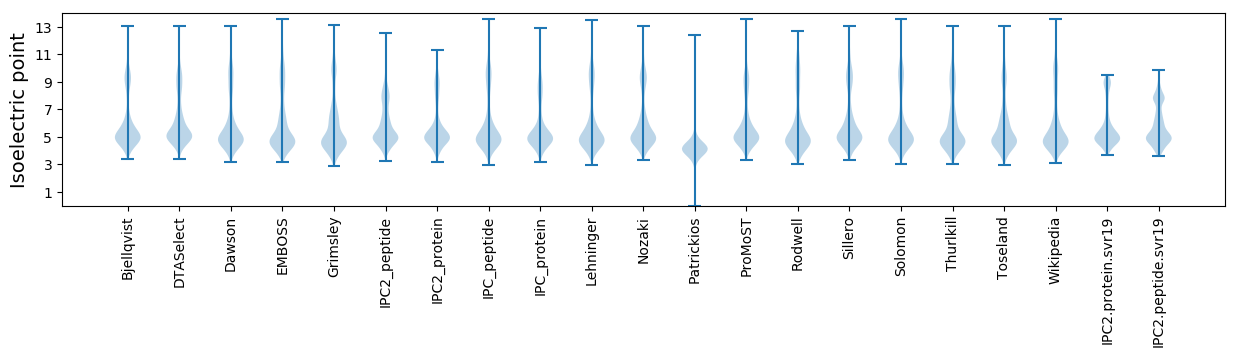
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A261G1F4|A0A261G1F4_9BIFI Sodium:proton antiporter OS=Bifidobacterium eulemuris OX=1765219 GN=BEUL_2004 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 4.82MKK15 pKa = 9.64HH16 pKa = 5.7GFRR19 pKa = 11.84LRR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84SGRR28 pKa = 11.84AVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.06GRR39 pKa = 11.84KK40 pKa = 8.87SLSAA44 pKa = 3.86
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 4.82MKK15 pKa = 9.64HH16 pKa = 5.7GFRR19 pKa = 11.84LRR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84SGRR28 pKa = 11.84AVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.06GRR39 pKa = 11.84KK40 pKa = 8.87SLSAA44 pKa = 3.86
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
844624 |
29 |
3136 |
363.1 |
39.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.154 ± 0.065 | 0.98 ± 0.018 |
6.76 ± 0.048 | 5.961 ± 0.046 |
3.488 ± 0.034 | 8.081 ± 0.04 |
1.99 ± 0.029 | 5.096 ± 0.043 |
3.288 ± 0.04 | 8.863 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.669 ± 0.025 | 3.202 ± 0.034 |
4.474 ± 0.034 | 3.263 ± 0.028 |
6.146 ± 0.056 | 6.15 ± 0.048 |
6.143 ± 0.045 | 7.963 ± 0.047 |
1.458 ± 0.021 | 2.866 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
