
Magnetospirillum magneticum (strain AMB-1 / ATCC 700264)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Magnetospirillum; Magnetospirillum magneticum
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
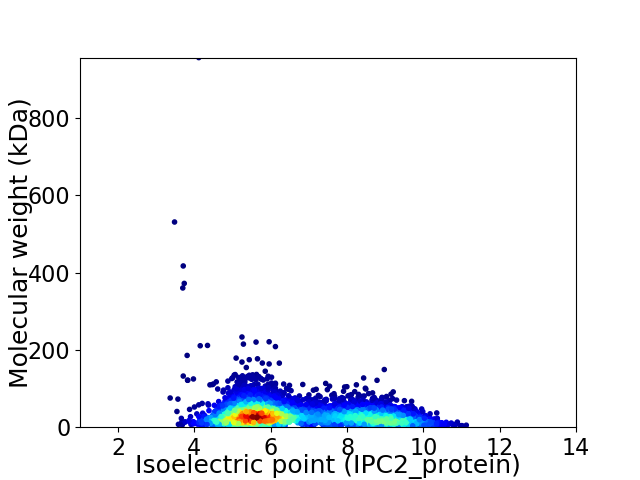
Virtual 2D-PAGE plot for 4515 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
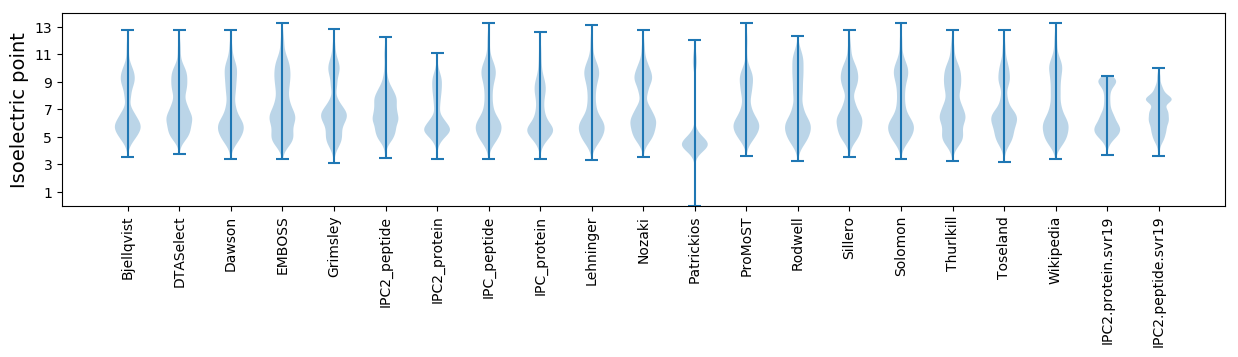
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q2W7X2|Q2W7X2_MAGSA Membrane-fusion protein OS=Magnetospirillum magneticum (strain AMB-1 / ATCC 700264) OX=342108 GN=amb1249 PE=3 SV=1
MM1 pKa = 6.87PTCRR5 pKa = 11.84PLGAGEE11 pKa = 4.75SMTKK15 pKa = 9.31TFAVASADD23 pKa = 3.34GTATQNVTVTINGANDD39 pKa = 3.15SATISGSTTGAVAEE53 pKa = 4.95DD54 pKa = 4.03GNGRR58 pKa = 11.84ATGTLSVADD67 pKa = 3.78VDD69 pKa = 4.11HH70 pKa = 6.53GQAHH74 pKa = 5.2VTAQTVQTDD83 pKa = 3.0QGTFSIGEE91 pKa = 3.92NGQWTFQVNSANADD105 pKa = 3.59VQALGAGEE113 pKa = 5.03SMTKK117 pKa = 9.31TFAVASADD125 pKa = 3.34GTATQNVSVTITGSNDD141 pKa = 2.67GPTVSGAVGLGHH153 pKa = 5.82TAEE156 pKa = 4.84DD157 pKa = 3.55NSVTFTKK164 pKa = 10.74AQLLANAHH172 pKa = 6.37DD173 pKa = 4.27TDD175 pKa = 3.57AHH177 pKa = 6.13DD178 pKa = 4.01TLSVSNLSAAHH189 pKa = 6.45GSIVDD194 pKa = 4.06NGNGTYY200 pKa = 10.21TFTPTEE206 pKa = 4.21TFSGHH211 pKa = 5.42VDD213 pKa = 3.07VSYY216 pKa = 9.91TVSDD220 pKa = 3.43GHH222 pKa = 6.81GGTTTGSAALDD233 pKa = 3.4VDD235 pKa = 4.42AVADD239 pKa = 4.18RR240 pKa = 11.84ATVSVAIGAPVEE252 pKa = 4.34TPNGSFTVTNLDD264 pKa = 3.52STASAGYY271 pKa = 9.45NNTYY275 pKa = 10.16GYY277 pKa = 11.44YY278 pKa = 11.27VMDD281 pKa = 5.13DD282 pKa = 3.56SGNPTSGGVIWSNVHH297 pKa = 6.46ASPGASVTISGVDD310 pKa = 3.46PDD312 pKa = 3.41RR313 pKa = 11.84VGFFLIPDD321 pKa = 4.4GGSQNSGLANGAALTFSKK339 pKa = 10.55DD340 pKa = 4.07ASGNWQASDD349 pKa = 3.41SAGHH353 pKa = 5.62VLSGAGTKK361 pKa = 10.27VLFDD365 pKa = 3.81KK366 pKa = 11.12SALNGDD372 pKa = 4.1HH373 pKa = 6.32MVHH376 pKa = 7.11SEE378 pKa = 3.96DD379 pKa = 3.51TSAVSGNQNWEE390 pKa = 4.1DD391 pKa = 3.52LTGGGDD397 pKa = 3.57RR398 pKa = 11.84DD399 pKa = 4.01YY400 pKa = 11.66NDD402 pKa = 3.5VNMSVTWGNAAPTATHH418 pKa = 7.28PLTVSASFPDD428 pKa = 3.44MDD430 pKa = 4.06GSEE433 pKa = 3.91GHH435 pKa = 6.33AVKK438 pKa = 10.6ISGLPSGSTLYY449 pKa = 10.81QNGVALVAGADD460 pKa = 3.72GSYY463 pKa = 10.96SLNPAQLGGLSVKK476 pKa = 9.82TPAGFNGDD484 pKa = 3.7LNVNVTAVATDD495 pKa = 3.82GTSVASSTASATVHH509 pKa = 7.41DD510 pKa = 5.39DD511 pKa = 3.89LSNHH515 pKa = 6.18GPDD518 pKa = 4.08AGDD521 pKa = 3.24ATSVHH526 pKa = 6.09GAMGATITGAAGVTDD541 pKa = 3.37SDD543 pKa = 4.35GDD545 pKa = 3.97HH546 pKa = 6.99LSYY549 pKa = 11.23AVGTGTSGPQHH560 pKa = 5.54GTVVVNTDD568 pKa = 2.8GTFTYY573 pKa = 9.57TPSGNYY579 pKa = 10.03SGTDD583 pKa = 3.1TFKK586 pKa = 9.75ITVSDD591 pKa = 3.67GHH593 pKa = 6.66GGTATEE599 pKa = 4.4TVNVTVEE606 pKa = 4.04NGAPTLTTANAATANDD622 pKa = 3.56HH623 pKa = 6.24SAVTGHH629 pKa = 5.31VTGSDD634 pKa = 3.55TPGDD638 pKa = 3.75TLSYY642 pKa = 11.13SLVDD646 pKa = 3.4ANGNHH651 pKa = 5.94VGSLVTDD658 pKa = 4.06HH659 pKa = 6.28GTVSINAATGDD670 pKa = 3.73YY671 pKa = 10.47TFTPNAANASLGVGASVTDD690 pKa = 3.56SFKK693 pKa = 11.3VVATDD698 pKa = 3.38NHH700 pKa = 6.33GADD703 pKa = 3.76SNLGTVAVTVTGSNDD718 pKa = 3.22GPVVTGVTGGTGNEE732 pKa = 4.66SITSARR738 pKa = 11.84SVVTGHH744 pKa = 6.88ISAGDD749 pKa = 3.48VDD751 pKa = 5.39AGDD754 pKa = 3.74TLSYY758 pKa = 11.11SVTDD762 pKa = 3.55NAAAGHH768 pKa = 6.56HH769 pKa = 5.97GTLAVNASGDD779 pKa = 3.82YY780 pKa = 10.58TFTASDD786 pKa = 4.32SNWHH790 pKa = 5.83GTDD793 pKa = 2.85SFTVQVSDD801 pKa = 3.5GHH803 pKa = 6.4GGTVNQTVAITVNAQADD820 pKa = 3.66AATINAQDD828 pKa = 3.77ASMAAQSGATFTASGGYY845 pKa = 10.17AEE847 pKa = 5.39GGSGSDD853 pKa = 3.88IINGSSANDD862 pKa = 3.38ILYY865 pKa = 10.8GDD867 pKa = 4.96DD868 pKa = 3.71PSGDD872 pKa = 3.76TEE874 pKa = 4.31VTVALNISAAAISGEE889 pKa = 4.35SVSSITLSNLPGGAVLNHH907 pKa = 5.9GHH909 pKa = 7.41DD910 pKa = 3.93NGDD913 pKa = 3.89GTWTLAPGDD922 pKa = 4.08LTGLTLTSNNGIGGTVDD939 pKa = 3.35VAVTTLDD946 pKa = 3.62GTSTAVTNSSFNVSFSGGFNDD967 pKa = 5.19TITGGVGADD976 pKa = 3.33TMYY979 pKa = 11.19GGAGNDD985 pKa = 3.43TFKK988 pKa = 11.29VSGASEE994 pKa = 4.43STGDD998 pKa = 3.69IYY1000 pKa = 11.39DD1001 pKa = 4.27GGSGTDD1007 pKa = 3.29TVLGSSGNDD1016 pKa = 3.33VISVTSNLGNMRR1028 pKa = 11.84SIEE1031 pKa = 4.27VIDD1034 pKa = 4.43GGAGTDD1040 pKa = 3.43TLLAGSGNDD1049 pKa = 3.36YY1050 pKa = 11.23LDD1052 pKa = 3.97FSGTTIRR1059 pKa = 11.84GVEE1062 pKa = 4.3VIDD1065 pKa = 3.54TGAGNDD1071 pKa = 4.06VITGSAGADD1080 pKa = 3.49TILTGTGDD1088 pKa = 3.81DD1089 pKa = 3.5TVVAIGGQTAGDD1101 pKa = 4.14VYY1103 pKa = 11.32DD1104 pKa = 5.0GGAGTDD1110 pKa = 3.59TLNVDD1115 pKa = 4.67LTPAQYY1121 pKa = 7.47TTAVRR1126 pKa = 11.84LEE1128 pKa = 4.15LMSFASFVDD1137 pKa = 3.85NPANAGKK1144 pKa = 10.47SFTFQSLGGLKK1155 pKa = 8.96ATNFEE1160 pKa = 4.41DD1161 pKa = 4.74LRR1163 pKa = 11.84VTVDD1167 pKa = 3.86GNDD1170 pKa = 2.86VDD1172 pKa = 5.78LNHH1175 pKa = 7.3PPQVTSVEE1183 pKa = 4.38SNYY1186 pKa = 9.72TAGHH1190 pKa = 6.63ADD1192 pKa = 3.71GVVHH1196 pKa = 6.57ATDD1199 pKa = 3.59SDD1201 pKa = 4.36GDD1203 pKa = 3.85TMSYY1207 pKa = 11.14SFGTNTNGTPITSMATAHH1225 pKa = 5.75GTVTIDD1231 pKa = 3.04SSTGEE1236 pKa = 4.04YY1237 pKa = 10.52HH1238 pKa = 6.04FTASDD1243 pKa = 3.71ANFKK1247 pKa = 9.65GTDD1250 pKa = 3.38TFSVTVKK1257 pKa = 10.67DD1258 pKa = 3.96GMGGTTTQKK1267 pKa = 10.72VNLDD1271 pKa = 3.83FNPGDD1276 pKa = 3.95DD1277 pKa = 3.69ATVVTGPTSLGGSDD1291 pKa = 4.99EE1292 pKa = 4.16DD1293 pKa = 3.78TQTFIRR1299 pKa = 11.84TDD1301 pKa = 3.29QLLANATDD1309 pKa = 3.57VDD1311 pKa = 4.21NTLHH1315 pKa = 5.81VANLSAVDD1323 pKa = 3.23SHH1325 pKa = 6.37GAVVGSFVHH1334 pKa = 6.91AVDD1337 pKa = 4.58ADD1339 pKa = 3.76GHH1341 pKa = 6.43DD1342 pKa = 4.31GYY1344 pKa = 11.68AFTPNANFAGDD1355 pKa = 3.39VTVNYY1360 pKa = 10.64DD1361 pKa = 3.14VVTDD1365 pKa = 3.58TGIATHH1371 pKa = 6.57TSGSLHH1377 pKa = 6.24VDD1379 pKa = 3.75AVADD1383 pKa = 3.66AATFDD1388 pKa = 3.74ATIGSNGVSVAHH1400 pKa = 7.05LGNDD1404 pKa = 3.35SKK1406 pKa = 11.66VVNLFVNDD1414 pKa = 4.05TSSGTAGFDD1423 pKa = 2.91IYY1425 pKa = 11.2LGGTNIGHH1433 pKa = 4.93YY1434 pKa = 9.09TVGRR1438 pKa = 11.84SYY1440 pKa = 11.51SSDD1443 pKa = 2.9KK1444 pKa = 11.1SVSLTLTDD1452 pKa = 3.76AQHH1455 pKa = 5.56TQLLNGADD1463 pKa = 3.42IKK1465 pKa = 11.05IVDD1468 pKa = 3.59NDD1470 pKa = 4.1GYY1472 pKa = 8.98NTRR1475 pKa = 11.84DD1476 pKa = 3.45VLVDD1480 pKa = 4.6RR1481 pKa = 11.84IQVDD1485 pKa = 3.55GLTFQAEE1492 pKa = 4.08NGILSGASTTSGYY1505 pKa = 11.01YY1506 pKa = 9.87GVSSEE1511 pKa = 4.43TYY1513 pKa = 10.14ARR1515 pKa = 11.84LNNVGSSVTFDD1526 pKa = 3.53FDD1528 pKa = 4.2SSVSHH1533 pKa = 6.56TSSGYY1538 pKa = 9.58HH1539 pKa = 6.06VDD1541 pKa = 4.55LAGHH1545 pKa = 7.09LNDD1548 pKa = 4.02TDD1550 pKa = 3.89GSEE1553 pKa = 4.15TLSYY1557 pKa = 10.54HH1558 pKa = 6.21IDD1560 pKa = 3.34AMTTGASLSYY1570 pKa = 9.79TGNEE1574 pKa = 3.68GTLVHH1579 pKa = 6.05NTDD1582 pKa = 3.66GSWDD1586 pKa = 3.83FNVDD1590 pKa = 2.78ASHH1593 pKa = 6.92YY1594 pKa = 10.62DD1595 pKa = 3.36GAVSLNLNVANGTAGFDD1612 pKa = 3.38VHH1614 pKa = 5.44VTANATEE1621 pKa = 4.09TSNLDD1626 pKa = 3.27VAGVGDD1632 pKa = 4.08TVHH1635 pKa = 7.28CDD1637 pKa = 3.27GAIGGAGAIPGITLVGNAGNDD1658 pKa = 4.06DD1659 pKa = 4.0IIGTNGNDD1667 pKa = 3.53LLLGGKK1673 pKa = 9.8GGATYY1678 pKa = 10.35RR1679 pKa = 11.84FSGGGCGSGGGWSIVQSDD1697 pKa = 4.1TNDD1700 pKa = 3.46VISAGAGDD1708 pKa = 3.87DD1709 pKa = 4.15VIYY1712 pKa = 11.07GDD1714 pKa = 3.94ARR1716 pKa = 11.84LVNGNIQITGSGNDD1730 pKa = 3.48VLDD1733 pKa = 4.34GGSGNDD1739 pKa = 3.86QIHH1742 pKa = 6.5GGAGNDD1748 pKa = 3.89TIIGGTGDD1756 pKa = 4.03DD1757 pKa = 5.02VMFGDD1762 pKa = 4.34QGNDD1766 pKa = 2.65TFLFDD1771 pKa = 4.72FGFGHH1776 pKa = 7.17DD1777 pKa = 3.76VVDD1780 pKa = 4.93GGRR1783 pKa = 11.84GSNWTDD1789 pKa = 3.21TLDD1792 pKa = 3.65LTHH1795 pKa = 7.57DD1796 pKa = 3.87NQISSVNIEE1805 pKa = 4.54GVSGWAVSVDD1815 pKa = 3.54AQGHH1819 pKa = 5.35HH1820 pKa = 6.2VAQATNGAHH1829 pKa = 7.07DD1830 pKa = 4.45ANGTIVVTNHH1840 pKa = 7.04DD1841 pKa = 4.03GSQDD1845 pKa = 3.32TIEE1848 pKa = 3.94FHH1850 pKa = 6.8NVEE1853 pKa = 4.2KK1854 pKa = 11.09VVWW1857 pKa = 3.61
MM1 pKa = 6.87PTCRR5 pKa = 11.84PLGAGEE11 pKa = 4.75SMTKK15 pKa = 9.31TFAVASADD23 pKa = 3.34GTATQNVTVTINGANDD39 pKa = 3.15SATISGSTTGAVAEE53 pKa = 4.95DD54 pKa = 4.03GNGRR58 pKa = 11.84ATGTLSVADD67 pKa = 3.78VDD69 pKa = 4.11HH70 pKa = 6.53GQAHH74 pKa = 5.2VTAQTVQTDD83 pKa = 3.0QGTFSIGEE91 pKa = 3.92NGQWTFQVNSANADD105 pKa = 3.59VQALGAGEE113 pKa = 5.03SMTKK117 pKa = 9.31TFAVASADD125 pKa = 3.34GTATQNVSVTITGSNDD141 pKa = 2.67GPTVSGAVGLGHH153 pKa = 5.82TAEE156 pKa = 4.84DD157 pKa = 3.55NSVTFTKK164 pKa = 10.74AQLLANAHH172 pKa = 6.37DD173 pKa = 4.27TDD175 pKa = 3.57AHH177 pKa = 6.13DD178 pKa = 4.01TLSVSNLSAAHH189 pKa = 6.45GSIVDD194 pKa = 4.06NGNGTYY200 pKa = 10.21TFTPTEE206 pKa = 4.21TFSGHH211 pKa = 5.42VDD213 pKa = 3.07VSYY216 pKa = 9.91TVSDD220 pKa = 3.43GHH222 pKa = 6.81GGTTTGSAALDD233 pKa = 3.4VDD235 pKa = 4.42AVADD239 pKa = 4.18RR240 pKa = 11.84ATVSVAIGAPVEE252 pKa = 4.34TPNGSFTVTNLDD264 pKa = 3.52STASAGYY271 pKa = 9.45NNTYY275 pKa = 10.16GYY277 pKa = 11.44YY278 pKa = 11.27VMDD281 pKa = 5.13DD282 pKa = 3.56SGNPTSGGVIWSNVHH297 pKa = 6.46ASPGASVTISGVDD310 pKa = 3.46PDD312 pKa = 3.41RR313 pKa = 11.84VGFFLIPDD321 pKa = 4.4GGSQNSGLANGAALTFSKK339 pKa = 10.55DD340 pKa = 4.07ASGNWQASDD349 pKa = 3.41SAGHH353 pKa = 5.62VLSGAGTKK361 pKa = 10.27VLFDD365 pKa = 3.81KK366 pKa = 11.12SALNGDD372 pKa = 4.1HH373 pKa = 6.32MVHH376 pKa = 7.11SEE378 pKa = 3.96DD379 pKa = 3.51TSAVSGNQNWEE390 pKa = 4.1DD391 pKa = 3.52LTGGGDD397 pKa = 3.57RR398 pKa = 11.84DD399 pKa = 4.01YY400 pKa = 11.66NDD402 pKa = 3.5VNMSVTWGNAAPTATHH418 pKa = 7.28PLTVSASFPDD428 pKa = 3.44MDD430 pKa = 4.06GSEE433 pKa = 3.91GHH435 pKa = 6.33AVKK438 pKa = 10.6ISGLPSGSTLYY449 pKa = 10.81QNGVALVAGADD460 pKa = 3.72GSYY463 pKa = 10.96SLNPAQLGGLSVKK476 pKa = 9.82TPAGFNGDD484 pKa = 3.7LNVNVTAVATDD495 pKa = 3.82GTSVASSTASATVHH509 pKa = 7.41DD510 pKa = 5.39DD511 pKa = 3.89LSNHH515 pKa = 6.18GPDD518 pKa = 4.08AGDD521 pKa = 3.24ATSVHH526 pKa = 6.09GAMGATITGAAGVTDD541 pKa = 3.37SDD543 pKa = 4.35GDD545 pKa = 3.97HH546 pKa = 6.99LSYY549 pKa = 11.23AVGTGTSGPQHH560 pKa = 5.54GTVVVNTDD568 pKa = 2.8GTFTYY573 pKa = 9.57TPSGNYY579 pKa = 10.03SGTDD583 pKa = 3.1TFKK586 pKa = 9.75ITVSDD591 pKa = 3.67GHH593 pKa = 6.66GGTATEE599 pKa = 4.4TVNVTVEE606 pKa = 4.04NGAPTLTTANAATANDD622 pKa = 3.56HH623 pKa = 6.24SAVTGHH629 pKa = 5.31VTGSDD634 pKa = 3.55TPGDD638 pKa = 3.75TLSYY642 pKa = 11.13SLVDD646 pKa = 3.4ANGNHH651 pKa = 5.94VGSLVTDD658 pKa = 4.06HH659 pKa = 6.28GTVSINAATGDD670 pKa = 3.73YY671 pKa = 10.47TFTPNAANASLGVGASVTDD690 pKa = 3.56SFKK693 pKa = 11.3VVATDD698 pKa = 3.38NHH700 pKa = 6.33GADD703 pKa = 3.76SNLGTVAVTVTGSNDD718 pKa = 3.22GPVVTGVTGGTGNEE732 pKa = 4.66SITSARR738 pKa = 11.84SVVTGHH744 pKa = 6.88ISAGDD749 pKa = 3.48VDD751 pKa = 5.39AGDD754 pKa = 3.74TLSYY758 pKa = 11.11SVTDD762 pKa = 3.55NAAAGHH768 pKa = 6.56HH769 pKa = 5.97GTLAVNASGDD779 pKa = 3.82YY780 pKa = 10.58TFTASDD786 pKa = 4.32SNWHH790 pKa = 5.83GTDD793 pKa = 2.85SFTVQVSDD801 pKa = 3.5GHH803 pKa = 6.4GGTVNQTVAITVNAQADD820 pKa = 3.66AATINAQDD828 pKa = 3.77ASMAAQSGATFTASGGYY845 pKa = 10.17AEE847 pKa = 5.39GGSGSDD853 pKa = 3.88IINGSSANDD862 pKa = 3.38ILYY865 pKa = 10.8GDD867 pKa = 4.96DD868 pKa = 3.71PSGDD872 pKa = 3.76TEE874 pKa = 4.31VTVALNISAAAISGEE889 pKa = 4.35SVSSITLSNLPGGAVLNHH907 pKa = 5.9GHH909 pKa = 7.41DD910 pKa = 3.93NGDD913 pKa = 3.89GTWTLAPGDD922 pKa = 4.08LTGLTLTSNNGIGGTVDD939 pKa = 3.35VAVTTLDD946 pKa = 3.62GTSTAVTNSSFNVSFSGGFNDD967 pKa = 5.19TITGGVGADD976 pKa = 3.33TMYY979 pKa = 11.19GGAGNDD985 pKa = 3.43TFKK988 pKa = 11.29VSGASEE994 pKa = 4.43STGDD998 pKa = 3.69IYY1000 pKa = 11.39DD1001 pKa = 4.27GGSGTDD1007 pKa = 3.29TVLGSSGNDD1016 pKa = 3.33VISVTSNLGNMRR1028 pKa = 11.84SIEE1031 pKa = 4.27VIDD1034 pKa = 4.43GGAGTDD1040 pKa = 3.43TLLAGSGNDD1049 pKa = 3.36YY1050 pKa = 11.23LDD1052 pKa = 3.97FSGTTIRR1059 pKa = 11.84GVEE1062 pKa = 4.3VIDD1065 pKa = 3.54TGAGNDD1071 pKa = 4.06VITGSAGADD1080 pKa = 3.49TILTGTGDD1088 pKa = 3.81DD1089 pKa = 3.5TVVAIGGQTAGDD1101 pKa = 4.14VYY1103 pKa = 11.32DD1104 pKa = 5.0GGAGTDD1110 pKa = 3.59TLNVDD1115 pKa = 4.67LTPAQYY1121 pKa = 7.47TTAVRR1126 pKa = 11.84LEE1128 pKa = 4.15LMSFASFVDD1137 pKa = 3.85NPANAGKK1144 pKa = 10.47SFTFQSLGGLKK1155 pKa = 8.96ATNFEE1160 pKa = 4.41DD1161 pKa = 4.74LRR1163 pKa = 11.84VTVDD1167 pKa = 3.86GNDD1170 pKa = 2.86VDD1172 pKa = 5.78LNHH1175 pKa = 7.3PPQVTSVEE1183 pKa = 4.38SNYY1186 pKa = 9.72TAGHH1190 pKa = 6.63ADD1192 pKa = 3.71GVVHH1196 pKa = 6.57ATDD1199 pKa = 3.59SDD1201 pKa = 4.36GDD1203 pKa = 3.85TMSYY1207 pKa = 11.14SFGTNTNGTPITSMATAHH1225 pKa = 5.75GTVTIDD1231 pKa = 3.04SSTGEE1236 pKa = 4.04YY1237 pKa = 10.52HH1238 pKa = 6.04FTASDD1243 pKa = 3.71ANFKK1247 pKa = 9.65GTDD1250 pKa = 3.38TFSVTVKK1257 pKa = 10.67DD1258 pKa = 3.96GMGGTTTQKK1267 pKa = 10.72VNLDD1271 pKa = 3.83FNPGDD1276 pKa = 3.95DD1277 pKa = 3.69ATVVTGPTSLGGSDD1291 pKa = 4.99EE1292 pKa = 4.16DD1293 pKa = 3.78TQTFIRR1299 pKa = 11.84TDD1301 pKa = 3.29QLLANATDD1309 pKa = 3.57VDD1311 pKa = 4.21NTLHH1315 pKa = 5.81VANLSAVDD1323 pKa = 3.23SHH1325 pKa = 6.37GAVVGSFVHH1334 pKa = 6.91AVDD1337 pKa = 4.58ADD1339 pKa = 3.76GHH1341 pKa = 6.43DD1342 pKa = 4.31GYY1344 pKa = 11.68AFTPNANFAGDD1355 pKa = 3.39VTVNYY1360 pKa = 10.64DD1361 pKa = 3.14VVTDD1365 pKa = 3.58TGIATHH1371 pKa = 6.57TSGSLHH1377 pKa = 6.24VDD1379 pKa = 3.75AVADD1383 pKa = 3.66AATFDD1388 pKa = 3.74ATIGSNGVSVAHH1400 pKa = 7.05LGNDD1404 pKa = 3.35SKK1406 pKa = 11.66VVNLFVNDD1414 pKa = 4.05TSSGTAGFDD1423 pKa = 2.91IYY1425 pKa = 11.2LGGTNIGHH1433 pKa = 4.93YY1434 pKa = 9.09TVGRR1438 pKa = 11.84SYY1440 pKa = 11.51SSDD1443 pKa = 2.9KK1444 pKa = 11.1SVSLTLTDD1452 pKa = 3.76AQHH1455 pKa = 5.56TQLLNGADD1463 pKa = 3.42IKK1465 pKa = 11.05IVDD1468 pKa = 3.59NDD1470 pKa = 4.1GYY1472 pKa = 8.98NTRR1475 pKa = 11.84DD1476 pKa = 3.45VLVDD1480 pKa = 4.6RR1481 pKa = 11.84IQVDD1485 pKa = 3.55GLTFQAEE1492 pKa = 4.08NGILSGASTTSGYY1505 pKa = 11.01YY1506 pKa = 9.87GVSSEE1511 pKa = 4.43TYY1513 pKa = 10.14ARR1515 pKa = 11.84LNNVGSSVTFDD1526 pKa = 3.53FDD1528 pKa = 4.2SSVSHH1533 pKa = 6.56TSSGYY1538 pKa = 9.58HH1539 pKa = 6.06VDD1541 pKa = 4.55LAGHH1545 pKa = 7.09LNDD1548 pKa = 4.02TDD1550 pKa = 3.89GSEE1553 pKa = 4.15TLSYY1557 pKa = 10.54HH1558 pKa = 6.21IDD1560 pKa = 3.34AMTTGASLSYY1570 pKa = 9.79TGNEE1574 pKa = 3.68GTLVHH1579 pKa = 6.05NTDD1582 pKa = 3.66GSWDD1586 pKa = 3.83FNVDD1590 pKa = 2.78ASHH1593 pKa = 6.92YY1594 pKa = 10.62DD1595 pKa = 3.36GAVSLNLNVANGTAGFDD1612 pKa = 3.38VHH1614 pKa = 5.44VTANATEE1621 pKa = 4.09TSNLDD1626 pKa = 3.27VAGVGDD1632 pKa = 4.08TVHH1635 pKa = 7.28CDD1637 pKa = 3.27GAIGGAGAIPGITLVGNAGNDD1658 pKa = 4.06DD1659 pKa = 4.0IIGTNGNDD1667 pKa = 3.53LLLGGKK1673 pKa = 9.8GGATYY1678 pKa = 10.35RR1679 pKa = 11.84FSGGGCGSGGGWSIVQSDD1697 pKa = 4.1TNDD1700 pKa = 3.46VISAGAGDD1708 pKa = 3.87DD1709 pKa = 4.15VIYY1712 pKa = 11.07GDD1714 pKa = 3.94ARR1716 pKa = 11.84LVNGNIQITGSGNDD1730 pKa = 3.48VLDD1733 pKa = 4.34GGSGNDD1739 pKa = 3.86QIHH1742 pKa = 6.5GGAGNDD1748 pKa = 3.89TIIGGTGDD1756 pKa = 4.03DD1757 pKa = 5.02VMFGDD1762 pKa = 4.34QGNDD1766 pKa = 2.65TFLFDD1771 pKa = 4.72FGFGHH1776 pKa = 7.17DD1777 pKa = 3.76VVDD1780 pKa = 4.93GGRR1783 pKa = 11.84GSNWTDD1789 pKa = 3.21TLDD1792 pKa = 3.65LTHH1795 pKa = 7.57DD1796 pKa = 3.87NQISSVNIEE1805 pKa = 4.54GVSGWAVSVDD1815 pKa = 3.54AQGHH1819 pKa = 5.35HH1820 pKa = 6.2VAQATNGAHH1829 pKa = 7.07DD1830 pKa = 4.45ANGTIVVTNHH1840 pKa = 7.04DD1841 pKa = 4.03GSQDD1845 pKa = 3.32TIEE1848 pKa = 3.94FHH1850 pKa = 6.8NVEE1853 pKa = 4.2KK1854 pKa = 11.09VVWW1857 pKa = 3.61
Molecular weight: 185.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q2W777|Q2W777_MAGSA Retron-type reverse transcriptase OS=Magnetospirillum magneticum (strain AMB-1 / ATCC 700264) OX=342108 GN=amb1494 PE=4 SV=1
MM1 pKa = 7.53IGALLRR7 pKa = 11.84RR8 pKa = 11.84LRR10 pKa = 11.84RR11 pKa = 11.84FFRR14 pKa = 11.84PPGSPVVIQSVRR26 pKa = 11.84GLSAPRR32 pKa = 11.84HH33 pKa = 4.87PQNN36 pKa = 3.88
MM1 pKa = 7.53IGALLRR7 pKa = 11.84RR8 pKa = 11.84LRR10 pKa = 11.84RR11 pKa = 11.84FFRR14 pKa = 11.84PPGSPVVIQSVRR26 pKa = 11.84GLSAPRR32 pKa = 11.84HH33 pKa = 4.87PQNN36 pKa = 3.88
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1451216 |
34 |
9529 |
321.4 |
34.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.644 ± 0.056 | 0.992 ± 0.016 |
5.671 ± 0.037 | 5.71 ± 0.05 |
3.392 ± 0.022 | 8.737 ± 0.054 |
2.135 ± 0.019 | 4.706 ± 0.027 |
3.51 ± 0.033 | 10.26 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.668 ± 0.02 | 2.489 ± 0.034 |
5.218 ± 0.049 | 3.093 ± 0.024 |
7.034 ± 0.07 | 5.468 ± 0.046 |
5.307 ± 0.089 | 7.613 ± 0.031 |
1.363 ± 0.016 | 1.99 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
