
Rosellinia necatrix mycoreovirus 3 (isolate W370) (RnMYRV-3) (Rosellinia anti-rot virus)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Mycoreovirus
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
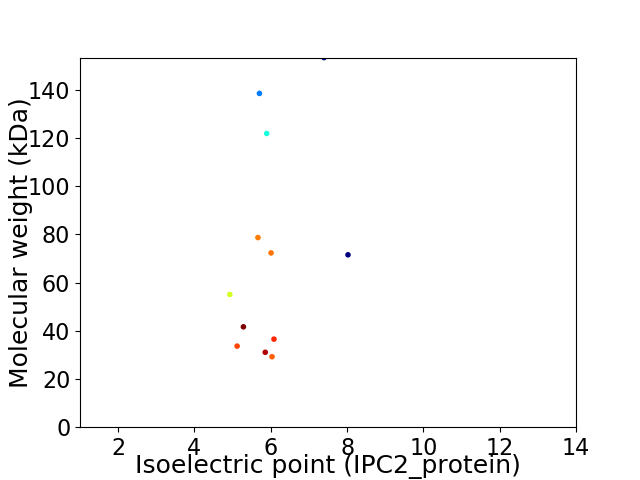
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8JXF4|Q8JXF4_MYRVW p6 OS=Rosellinia necatrix mycoreovirus 3 (isolate W370) OX=311229 PE=4 SV=1
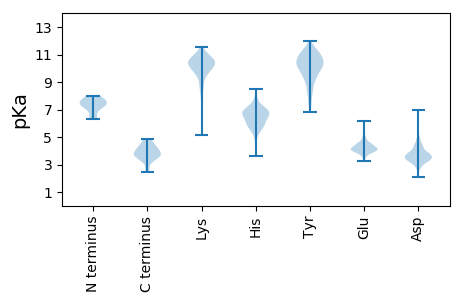
MM1 pKa = 7.29AQQVHH6 pKa = 6.28AVQLSPRR13 pKa = 11.84LRR15 pKa = 11.84NDD17 pKa = 2.91VLLQLTKK24 pKa = 10.5RR25 pKa = 11.84SLMTIDD31 pKa = 3.73TGTILQQVEE40 pKa = 4.29LPLNLFLLGVCFVGDD55 pKa = 3.45GDD57 pKa = 4.43VPYY60 pKa = 11.28LLIRR64 pKa = 11.84QIAGYY69 pKa = 7.09TWHH72 pKa = 7.62IDD74 pKa = 3.14PGYY77 pKa = 9.45GIEE80 pKa = 4.13MYY82 pKa = 10.21EE83 pKa = 3.91VDD85 pKa = 4.33VGEE88 pKa = 4.46EE89 pKa = 4.13MPVLQARR96 pKa = 11.84IDD98 pKa = 3.63SLAYY102 pKa = 10.85VMTIDD107 pKa = 4.29EE108 pKa = 4.72RR109 pKa = 11.84DD110 pKa = 3.38YY111 pKa = 10.54LYY113 pKa = 10.35PVITVDD119 pKa = 3.84EE120 pKa = 4.91DD121 pKa = 3.66GRR123 pKa = 11.84TLNINYY129 pKa = 9.12TDD131 pKa = 4.31DD132 pKa = 3.91PSLRR136 pKa = 11.84HH137 pKa = 6.79DD138 pKa = 3.44VDD140 pKa = 3.61TYY142 pKa = 11.73ARR144 pKa = 11.84LSRR147 pKa = 11.84EE148 pKa = 3.99LQLVRR153 pKa = 11.84DD154 pKa = 4.86DD155 pKa = 3.65ISVRR159 pKa = 11.84SHH161 pKa = 6.77DD162 pKa = 4.49RR163 pKa = 11.84RR164 pKa = 11.84PDD166 pKa = 3.37SPSIYY171 pKa = 9.26SDD173 pKa = 2.97QRR175 pKa = 11.84SRR177 pKa = 11.84PNTPPRR183 pKa = 11.84PITPPYY189 pKa = 9.94LRR191 pKa = 11.84DD192 pKa = 3.45YY193 pKa = 11.03DD194 pKa = 4.15VEE196 pKa = 4.77SEE198 pKa = 4.96DD199 pKa = 3.6DD200 pKa = 3.82TEE202 pKa = 5.83DD203 pKa = 3.53GFVRR207 pKa = 11.84ISYY210 pKa = 10.56ASLRR214 pKa = 11.84AAYY217 pKa = 9.24RR218 pKa = 11.84PPNTTTTLVRR228 pKa = 11.84SMLYY232 pKa = 10.42NPIPVPPPQPLEE244 pKa = 4.31TRR246 pKa = 11.84WSTFYY251 pKa = 11.62AMDD254 pKa = 4.62ADD256 pKa = 3.91LRR258 pKa = 11.84HH259 pKa = 6.74KK260 pKa = 10.23IRR262 pKa = 11.84HH263 pKa = 5.37SLNFPLSHH271 pKa = 6.53GEE273 pKa = 3.56WRR275 pKa = 11.84LSDD278 pKa = 3.33NALRR282 pKa = 11.84GVGMMPRR289 pKa = 11.84PSGGVYY295 pKa = 7.14QHH297 pKa = 6.93ISHH300 pKa = 6.8MCNDD304 pKa = 3.71PEE306 pKa = 4.7SRR308 pKa = 11.84LLQSTEE314 pKa = 4.05WEE316 pKa = 4.05HH317 pKa = 6.14MIYY320 pKa = 9.94MIDD323 pKa = 3.78KK324 pKa = 10.41RR325 pKa = 11.84YY326 pKa = 9.62IDD328 pKa = 4.23LGAVCSNGLGATVLSIDD345 pKa = 3.63AATSLEE351 pKa = 3.98MAKK354 pKa = 10.52DD355 pKa = 3.44VLYY358 pKa = 11.03LFGRR362 pKa = 11.84AVVTFRR368 pKa = 11.84EE369 pKa = 4.82WKK371 pKa = 10.47RR372 pKa = 11.84EE373 pKa = 3.86TADD376 pKa = 3.52EE377 pKa = 4.41NEE379 pKa = 3.82LLMRR383 pKa = 11.84VFPDD387 pKa = 3.83KK388 pKa = 10.75IITVVDD394 pKa = 3.31YY395 pKa = 11.29SYY397 pKa = 11.92LNTFKK402 pKa = 11.03YY403 pKa = 10.51RR404 pKa = 11.84IDD406 pKa = 3.21QRR408 pKa = 11.84EE409 pKa = 4.05NVRR412 pKa = 11.84MVLGLINGALSDD424 pKa = 4.02SNRR427 pKa = 11.84EE428 pKa = 3.35EE429 pKa = 4.2WASVLALIYY438 pKa = 10.0MVTNRR443 pKa = 11.84QLPGSQDD450 pKa = 3.1AGVDD454 pKa = 4.01DD455 pKa = 3.56IHH457 pKa = 8.47VLLRR461 pKa = 11.84PPEE464 pKa = 4.65SGLSYY469 pKa = 10.98LVLLALIIVVSDD481 pKa = 3.44RR482 pKa = 3.67
MM1 pKa = 7.29AQQVHH6 pKa = 6.28AVQLSPRR13 pKa = 11.84LRR15 pKa = 11.84NDD17 pKa = 2.91VLLQLTKK24 pKa = 10.5RR25 pKa = 11.84SLMTIDD31 pKa = 3.73TGTILQQVEE40 pKa = 4.29LPLNLFLLGVCFVGDD55 pKa = 3.45GDD57 pKa = 4.43VPYY60 pKa = 11.28LLIRR64 pKa = 11.84QIAGYY69 pKa = 7.09TWHH72 pKa = 7.62IDD74 pKa = 3.14PGYY77 pKa = 9.45GIEE80 pKa = 4.13MYY82 pKa = 10.21EE83 pKa = 3.91VDD85 pKa = 4.33VGEE88 pKa = 4.46EE89 pKa = 4.13MPVLQARR96 pKa = 11.84IDD98 pKa = 3.63SLAYY102 pKa = 10.85VMTIDD107 pKa = 4.29EE108 pKa = 4.72RR109 pKa = 11.84DD110 pKa = 3.38YY111 pKa = 10.54LYY113 pKa = 10.35PVITVDD119 pKa = 3.84EE120 pKa = 4.91DD121 pKa = 3.66GRR123 pKa = 11.84TLNINYY129 pKa = 9.12TDD131 pKa = 4.31DD132 pKa = 3.91PSLRR136 pKa = 11.84HH137 pKa = 6.79DD138 pKa = 3.44VDD140 pKa = 3.61TYY142 pKa = 11.73ARR144 pKa = 11.84LSRR147 pKa = 11.84EE148 pKa = 3.99LQLVRR153 pKa = 11.84DD154 pKa = 4.86DD155 pKa = 3.65ISVRR159 pKa = 11.84SHH161 pKa = 6.77DD162 pKa = 4.49RR163 pKa = 11.84RR164 pKa = 11.84PDD166 pKa = 3.37SPSIYY171 pKa = 9.26SDD173 pKa = 2.97QRR175 pKa = 11.84SRR177 pKa = 11.84PNTPPRR183 pKa = 11.84PITPPYY189 pKa = 9.94LRR191 pKa = 11.84DD192 pKa = 3.45YY193 pKa = 11.03DD194 pKa = 4.15VEE196 pKa = 4.77SEE198 pKa = 4.96DD199 pKa = 3.6DD200 pKa = 3.82TEE202 pKa = 5.83DD203 pKa = 3.53GFVRR207 pKa = 11.84ISYY210 pKa = 10.56ASLRR214 pKa = 11.84AAYY217 pKa = 9.24RR218 pKa = 11.84PPNTTTTLVRR228 pKa = 11.84SMLYY232 pKa = 10.42NPIPVPPPQPLEE244 pKa = 4.31TRR246 pKa = 11.84WSTFYY251 pKa = 11.62AMDD254 pKa = 4.62ADD256 pKa = 3.91LRR258 pKa = 11.84HH259 pKa = 6.74KK260 pKa = 10.23IRR262 pKa = 11.84HH263 pKa = 5.37SLNFPLSHH271 pKa = 6.53GEE273 pKa = 3.56WRR275 pKa = 11.84LSDD278 pKa = 3.33NALRR282 pKa = 11.84GVGMMPRR289 pKa = 11.84PSGGVYY295 pKa = 7.14QHH297 pKa = 6.93ISHH300 pKa = 6.8MCNDD304 pKa = 3.71PEE306 pKa = 4.7SRR308 pKa = 11.84LLQSTEE314 pKa = 4.05WEE316 pKa = 4.05HH317 pKa = 6.14MIYY320 pKa = 9.94MIDD323 pKa = 3.78KK324 pKa = 10.41RR325 pKa = 11.84YY326 pKa = 9.62IDD328 pKa = 4.23LGAVCSNGLGATVLSIDD345 pKa = 3.63AATSLEE351 pKa = 3.98MAKK354 pKa = 10.52DD355 pKa = 3.44VLYY358 pKa = 11.03LFGRR362 pKa = 11.84AVVTFRR368 pKa = 11.84EE369 pKa = 4.82WKK371 pKa = 10.47RR372 pKa = 11.84EE373 pKa = 3.86TADD376 pKa = 3.52EE377 pKa = 4.41NEE379 pKa = 3.82LLMRR383 pKa = 11.84VFPDD387 pKa = 3.83KK388 pKa = 10.75IITVVDD394 pKa = 3.31YY395 pKa = 11.29SYY397 pKa = 11.92LNTFKK402 pKa = 11.03YY403 pKa = 10.51RR404 pKa = 11.84IDD406 pKa = 3.21QRR408 pKa = 11.84EE409 pKa = 4.05NVRR412 pKa = 11.84MVLGLINGALSDD424 pKa = 4.02SNRR427 pKa = 11.84EE428 pKa = 3.35EE429 pKa = 4.2WASVLALIYY438 pKa = 10.0MVTNRR443 pKa = 11.84QLPGSQDD450 pKa = 3.1AGVDD454 pKa = 4.01DD455 pKa = 3.56IHH457 pKa = 8.47VLLRR461 pKa = 11.84PPEE464 pKa = 4.65SGLSYY469 pKa = 10.98LVLLALIIVVSDD481 pKa = 3.44RR482 pKa = 3.67
Molecular weight: 55.07 kDa
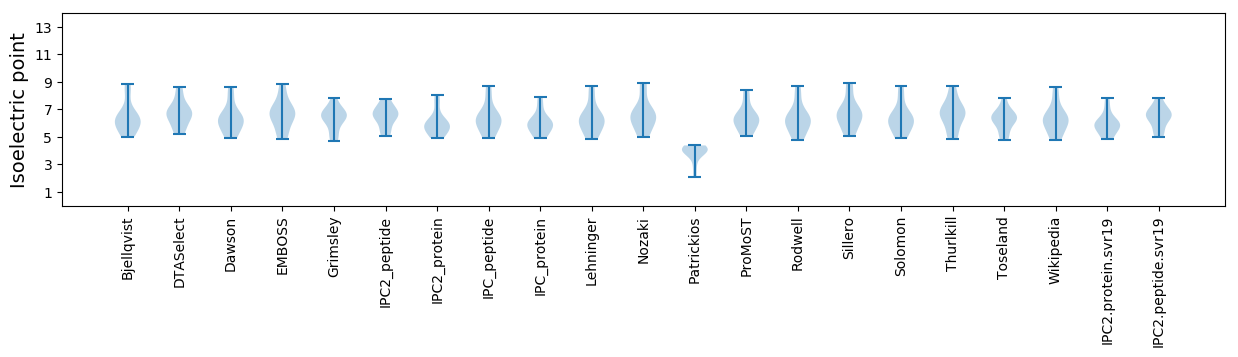
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8JXF5|Q8JXF5_MYRVW PX OS=Rosellinia necatrix mycoreovirus 3 (isolate W370) OX=311229 PE=4 SV=1
MM1 pKa = 7.13QHH3 pKa = 5.85MSHH6 pKa = 5.24YY7 pKa = 9.9HH8 pKa = 5.41FRR10 pKa = 11.84LICTGPYY17 pKa = 9.66AVLRR21 pKa = 11.84IILNDD26 pKa = 3.42CTPPMLHH33 pKa = 5.94GTLMSSQTTGKK44 pKa = 8.74PQYY47 pKa = 10.36LRR49 pKa = 11.84AVPHH53 pKa = 6.6IFMIDD58 pKa = 3.34SMDD61 pKa = 3.55KK62 pKa = 10.78FSTSHH67 pKa = 5.96ATLLSKK73 pKa = 10.8CPQEE77 pKa = 4.27YY78 pKa = 9.89VSSDD82 pKa = 2.66ITVITSPTAQDD93 pKa = 2.93IARR96 pKa = 11.84SIRR99 pKa = 11.84ANKK102 pKa = 9.97HH103 pKa = 3.99VVCATLPVGVFNDD116 pKa = 4.7AIVFSSITVPDD127 pKa = 3.4CTKK130 pKa = 9.05YY131 pKa = 10.77LRR133 pKa = 11.84RR134 pKa = 11.84TFHH137 pKa = 6.63ALLRR141 pKa = 11.84RR142 pKa = 11.84LSFGHH147 pKa = 5.88SCYY150 pKa = 10.57SCSNVLQLSLPPWLSVLASGWKK172 pKa = 9.74RR173 pKa = 11.84YY174 pKa = 9.44VPSSSNDD181 pKa = 3.45DD182 pKa = 3.4LAKK185 pKa = 10.43FYY187 pKa = 10.02LQPSSSPNQIVTNVQDD203 pKa = 3.81LQDD206 pKa = 3.69FVTNQLKK213 pKa = 10.57SDD215 pKa = 3.53VDD217 pKa = 3.66VVYY220 pKa = 10.68FDD222 pKa = 6.22DD223 pKa = 4.77VLTTDD228 pKa = 5.41GITIGEE234 pKa = 4.54KK235 pKa = 9.85IRR237 pKa = 11.84STGAQFNVARR247 pKa = 11.84DD248 pKa = 3.66LTHH251 pKa = 7.06FNIIKK256 pKa = 9.53QRR258 pKa = 11.84YY259 pKa = 7.6VEE261 pKa = 4.02QTSIRR266 pKa = 11.84TRR268 pKa = 11.84RR269 pKa = 11.84QHH271 pKa = 6.93KK272 pKa = 10.13KK273 pKa = 10.03DD274 pKa = 3.16LHH276 pKa = 5.43ASKK279 pKa = 10.6RR280 pKa = 11.84KK281 pKa = 6.23QTITARR287 pKa = 11.84VFPPVYY293 pKa = 10.39LFDD296 pKa = 3.19ITVTIKK302 pKa = 9.95SWSSAMPMSQQRR314 pKa = 11.84RR315 pKa = 11.84LSRR318 pKa = 11.84VYY320 pKa = 10.91SDD322 pKa = 5.91LIQLSKK328 pKa = 11.15KK329 pKa = 9.65PLTVLHH335 pKa = 5.94MLSWDD340 pKa = 3.83GPNTISTDD348 pKa = 3.57DD349 pKa = 5.2DD350 pKa = 3.08IWSQFTEE357 pKa = 3.93WLFGVVQNVDD367 pKa = 2.81EE368 pKa = 4.44TMARR372 pKa = 11.84LLTAIPYY379 pKa = 7.7MSVMVAVSRR388 pKa = 11.84RR389 pKa = 11.84GFGTWIDD396 pKa = 3.69FRR398 pKa = 11.84NWTEE402 pKa = 4.07GFSLSKK408 pKa = 10.41ILVGRR413 pKa = 11.84TYY415 pKa = 10.47MVGKK419 pKa = 9.93KK420 pKa = 10.13NSGKK424 pKa = 10.94GMIGKK429 pKa = 8.59LIWKK433 pKa = 9.44LGVPVIDD440 pKa = 3.52SDD442 pKa = 4.16DD443 pKa = 3.68YY444 pKa = 12.01GRR446 pKa = 11.84VLLIAEE452 pKa = 4.36DD453 pKa = 3.35QGVPLNEE460 pKa = 3.57AVHH463 pKa = 5.98RR464 pKa = 11.84HH465 pKa = 5.36FSLTYY470 pKa = 7.42QQRR473 pKa = 11.84DD474 pKa = 3.28TAPTVFEE481 pKa = 4.58KK482 pKa = 11.1AMDD485 pKa = 5.14DD486 pKa = 3.39IVRR489 pKa = 11.84DD490 pKa = 3.73LAVVRR495 pKa = 11.84YY496 pKa = 9.03AIPEE500 pKa = 3.9VDD502 pKa = 4.14HH503 pKa = 6.97PALIAFGRR511 pKa = 11.84VYY513 pKa = 11.21DD514 pKa = 4.32EE515 pKa = 4.27LTSKK519 pKa = 10.83YY520 pKa = 10.3KK521 pKa = 10.33YY522 pKa = 10.47ADD524 pKa = 3.27FEE526 pKa = 4.53RR527 pKa = 11.84VVRR530 pKa = 11.84SHH532 pKa = 6.64IASEE536 pKa = 4.63GVLAPDD542 pKa = 3.56GTLLKK547 pKa = 10.57VDD549 pKa = 3.85DD550 pKa = 4.35RR551 pKa = 11.84FVFSTHH557 pKa = 6.2CSEE560 pKa = 5.02EE561 pKa = 3.95ATQVLGANYY570 pKa = 8.8MFQLSTAIDD579 pKa = 3.77SYY581 pKa = 11.36IGVLLRR587 pKa = 11.84GQHH590 pKa = 5.87SNAISEE596 pKa = 4.48LMLAVYY602 pKa = 10.0YY603 pKa = 10.69DD604 pKa = 4.57RR605 pKa = 11.84IHH607 pKa = 7.83VNIFDD612 pKa = 4.57LVPTGVVCTVLRR624 pKa = 11.84TGLARR629 pKa = 11.84LVPKK633 pKa = 10.64AA634 pKa = 3.7
MM1 pKa = 7.13QHH3 pKa = 5.85MSHH6 pKa = 5.24YY7 pKa = 9.9HH8 pKa = 5.41FRR10 pKa = 11.84LICTGPYY17 pKa = 9.66AVLRR21 pKa = 11.84IILNDD26 pKa = 3.42CTPPMLHH33 pKa = 5.94GTLMSSQTTGKK44 pKa = 8.74PQYY47 pKa = 10.36LRR49 pKa = 11.84AVPHH53 pKa = 6.6IFMIDD58 pKa = 3.34SMDD61 pKa = 3.55KK62 pKa = 10.78FSTSHH67 pKa = 5.96ATLLSKK73 pKa = 10.8CPQEE77 pKa = 4.27YY78 pKa = 9.89VSSDD82 pKa = 2.66ITVITSPTAQDD93 pKa = 2.93IARR96 pKa = 11.84SIRR99 pKa = 11.84ANKK102 pKa = 9.97HH103 pKa = 3.99VVCATLPVGVFNDD116 pKa = 4.7AIVFSSITVPDD127 pKa = 3.4CTKK130 pKa = 9.05YY131 pKa = 10.77LRR133 pKa = 11.84RR134 pKa = 11.84TFHH137 pKa = 6.63ALLRR141 pKa = 11.84RR142 pKa = 11.84LSFGHH147 pKa = 5.88SCYY150 pKa = 10.57SCSNVLQLSLPPWLSVLASGWKK172 pKa = 9.74RR173 pKa = 11.84YY174 pKa = 9.44VPSSSNDD181 pKa = 3.45DD182 pKa = 3.4LAKK185 pKa = 10.43FYY187 pKa = 10.02LQPSSSPNQIVTNVQDD203 pKa = 3.81LQDD206 pKa = 3.69FVTNQLKK213 pKa = 10.57SDD215 pKa = 3.53VDD217 pKa = 3.66VVYY220 pKa = 10.68FDD222 pKa = 6.22DD223 pKa = 4.77VLTTDD228 pKa = 5.41GITIGEE234 pKa = 4.54KK235 pKa = 9.85IRR237 pKa = 11.84STGAQFNVARR247 pKa = 11.84DD248 pKa = 3.66LTHH251 pKa = 7.06FNIIKK256 pKa = 9.53QRR258 pKa = 11.84YY259 pKa = 7.6VEE261 pKa = 4.02QTSIRR266 pKa = 11.84TRR268 pKa = 11.84RR269 pKa = 11.84QHH271 pKa = 6.93KK272 pKa = 10.13KK273 pKa = 10.03DD274 pKa = 3.16LHH276 pKa = 5.43ASKK279 pKa = 10.6RR280 pKa = 11.84KK281 pKa = 6.23QTITARR287 pKa = 11.84VFPPVYY293 pKa = 10.39LFDD296 pKa = 3.19ITVTIKK302 pKa = 9.95SWSSAMPMSQQRR314 pKa = 11.84RR315 pKa = 11.84LSRR318 pKa = 11.84VYY320 pKa = 10.91SDD322 pKa = 5.91LIQLSKK328 pKa = 11.15KK329 pKa = 9.65PLTVLHH335 pKa = 5.94MLSWDD340 pKa = 3.83GPNTISTDD348 pKa = 3.57DD349 pKa = 5.2DD350 pKa = 3.08IWSQFTEE357 pKa = 3.93WLFGVVQNVDD367 pKa = 2.81EE368 pKa = 4.44TMARR372 pKa = 11.84LLTAIPYY379 pKa = 7.7MSVMVAVSRR388 pKa = 11.84RR389 pKa = 11.84GFGTWIDD396 pKa = 3.69FRR398 pKa = 11.84NWTEE402 pKa = 4.07GFSLSKK408 pKa = 10.41ILVGRR413 pKa = 11.84TYY415 pKa = 10.47MVGKK419 pKa = 9.93KK420 pKa = 10.13NSGKK424 pKa = 10.94GMIGKK429 pKa = 8.59LIWKK433 pKa = 9.44LGVPVIDD440 pKa = 3.52SDD442 pKa = 4.16DD443 pKa = 3.68YY444 pKa = 12.01GRR446 pKa = 11.84VLLIAEE452 pKa = 4.36DD453 pKa = 3.35QGVPLNEE460 pKa = 3.57AVHH463 pKa = 5.98RR464 pKa = 11.84HH465 pKa = 5.36FSLTYY470 pKa = 7.42QQRR473 pKa = 11.84DD474 pKa = 3.28TAPTVFEE481 pKa = 4.58KK482 pKa = 11.1AMDD485 pKa = 5.14DD486 pKa = 3.39IVRR489 pKa = 11.84DD490 pKa = 3.73LAVVRR495 pKa = 11.84YY496 pKa = 9.03AIPEE500 pKa = 3.9VDD502 pKa = 4.14HH503 pKa = 6.97PALIAFGRR511 pKa = 11.84VYY513 pKa = 11.21DD514 pKa = 4.32EE515 pKa = 4.27LTSKK519 pKa = 10.83YY520 pKa = 10.3KK521 pKa = 10.33YY522 pKa = 10.47ADD524 pKa = 3.27FEE526 pKa = 4.53RR527 pKa = 11.84VVRR530 pKa = 11.84SHH532 pKa = 6.64IASEE536 pKa = 4.63GVLAPDD542 pKa = 3.56GTLLKK547 pKa = 10.57VDD549 pKa = 3.85DD550 pKa = 4.35RR551 pKa = 11.84FVFSTHH557 pKa = 6.2CSEE560 pKa = 5.02EE561 pKa = 3.95ATQVLGANYY570 pKa = 8.8MFQLSTAIDD579 pKa = 3.77SYY581 pKa = 11.36IGVLLRR587 pKa = 11.84GQHH590 pKa = 5.87SNAISEE596 pKa = 4.48LMLAVYY602 pKa = 10.0YY603 pKa = 10.69DD604 pKa = 4.57RR605 pKa = 11.84IHH607 pKa = 7.83VNIFDD612 pKa = 4.57LVPTGVVCTVLRR624 pKa = 11.84TGLARR629 pKa = 11.84LVPKK633 pKa = 10.64AA634 pKa = 3.7
Molecular weight: 71.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7721 |
265 |
1360 |
643.4 |
71.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.696 ± 0.282 | 1.567 ± 0.116 |
6.282 ± 0.268 | 4.352 ± 0.256 |
4.287 ± 0.256 | 6.178 ± 0.34 |
2.953 ± 0.219 | 5.453 ± 0.186 |
2.862 ± 0.281 | 8.937 ± 0.312 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.875 ± 0.14 | 3.536 ± 0.267 |
5.44 ± 0.226 | 3.393 ± 0.205 |
6.774 ± 0.389 | 8.432 ± 0.504 |
6.567 ± 0.244 | 8.483 ± 0.376 |
1.295 ± 0.113 | 3.639 ± 0.232 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
