
Shahe tombus-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.87
Get precalculated fractions of proteins
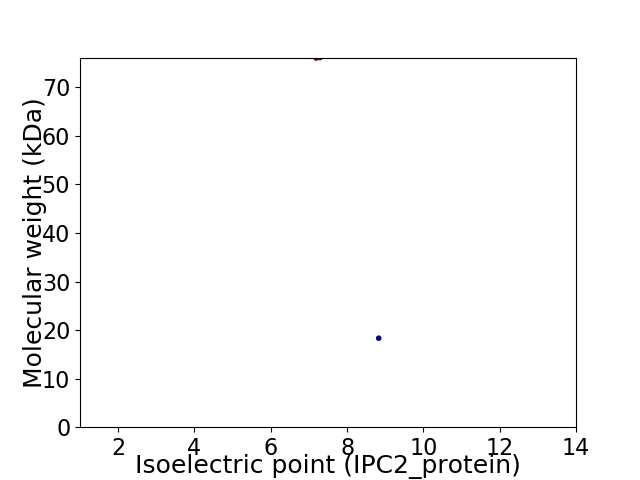
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KG67|A0A1L3KG67_9VIRU Uncharacterized protein OS=Shahe tombus-like virus 2 OX=1923456 PE=4 SV=1
MM1 pKa = 7.25EE2 pKa = 5.14QFICSYY8 pKa = 10.65KK9 pKa = 10.14GAKK12 pKa = 7.95QHH14 pKa = 5.24TVRR17 pKa = 11.84AAAEE21 pKa = 4.0DD22 pKa = 4.25VLRR25 pKa = 11.84HH26 pKa = 4.98PAVRR30 pKa = 11.84RR31 pKa = 11.84DD32 pKa = 3.53AKK34 pKa = 10.95LKK36 pKa = 9.82TFVKK40 pKa = 10.52AEE42 pKa = 3.94KK43 pKa = 10.48LNLSSKK49 pKa = 9.85PDD51 pKa = 3.65PAPRR55 pKa = 11.84VIQPRR60 pKa = 11.84DD61 pKa = 3.16VRR63 pKa = 11.84YY64 pKa = 8.53NCEE67 pKa = 3.54VGPYY71 pKa = 9.88LKK73 pKa = 10.6AHH75 pKa = 6.01EE76 pKa = 4.09HH77 pKa = 5.56TIYY80 pKa = 10.67RR81 pKa = 11.84GIAKK85 pKa = 9.48IWGGPTVMKK94 pKa = 10.53GFNSVDD100 pKa = 3.38TAKK103 pKa = 10.5HH104 pKa = 4.72MRR106 pKa = 11.84EE107 pKa = 4.02MWDD110 pKa = 4.38SIHH113 pKa = 7.11DD114 pKa = 3.73PCGIGLDD121 pKa = 3.45ASRR124 pKa = 11.84FDD126 pKa = 3.33QHH128 pKa = 7.84VSEE131 pKa = 4.65EE132 pKa = 4.04ALRR135 pKa = 11.84WEE137 pKa = 4.0HH138 pKa = 6.29SVYY141 pKa = 11.32NGMFKK146 pKa = 10.46SKK148 pKa = 10.46RR149 pKa = 11.84LAEE152 pKa = 4.12LLTWQINNVGTSYY165 pKa = 10.32TPEE168 pKa = 3.56GRR170 pKa = 11.84VTYY173 pKa = 9.83KK174 pKa = 10.97VKK176 pKa = 9.91GCRR179 pKa = 11.84MSGDD183 pKa = 3.68MNTSLGNCLIMCGMVWEE200 pKa = 5.2LCQQLGCPARR210 pKa = 11.84LANNGDD216 pKa = 3.82DD217 pKa = 4.47CMLIVPRR224 pKa = 11.84SWEE227 pKa = 3.43QRR229 pKa = 11.84VRR231 pKa = 11.84DD232 pKa = 4.7KK233 pKa = 10.83IASWFLDD240 pKa = 3.47FGFTMKK246 pKa = 10.55VEE248 pKa = 3.99DD249 pKa = 3.54TVYY252 pKa = 10.43EE253 pKa = 4.07FEE255 pKa = 5.08QIEE258 pKa = 4.63FCQTRR263 pKa = 11.84PVFASNRR270 pKa = 11.84WVMTRR275 pKa = 11.84DD276 pKa = 3.27YY277 pKa = 10.65RR278 pKa = 11.84VVMDD282 pKa = 4.99KK283 pKa = 10.58DD284 pKa = 4.19TFCLHH289 pKa = 7.44PDD291 pKa = 3.06NMPYY295 pKa = 10.22EE296 pKa = 3.97RR297 pKa = 11.84WLRR300 pKa = 11.84GVGTAGLALASGVPVLQTFYY320 pKa = 10.0TQLVALGGGQGEE332 pKa = 5.01LIDD335 pKa = 4.33GSGMSYY341 pKa = 8.8MAKK344 pKa = 10.39GLTPDD349 pKa = 4.06ASPITTEE356 pKa = 4.1ARR358 pKa = 11.84VSFYY362 pKa = 10.79KK363 pKa = 10.79AFGMPPWVQEE373 pKa = 4.03AYY375 pKa = 9.99EE376 pKa = 4.08EE377 pKa = 4.61EE378 pKa = 4.48IVASASSTNLGLIGSRR394 pKa = 11.84YY395 pKa = 7.53TYY397 pKa = 10.88HH398 pKa = 6.89NNTLAFQGMTGKK410 pKa = 8.88MKK412 pKa = 10.62KK413 pKa = 9.98PSTQKK418 pKa = 11.24SMIRR422 pKa = 11.84NIPIDD427 pKa = 3.92VPGGPPGYY435 pKa = 9.5TNFQRR440 pKa = 11.84KK441 pKa = 6.43TGPRR445 pKa = 11.84VTATKK450 pKa = 10.6AGLQIANSEE459 pKa = 4.08QFLPLSADD467 pKa = 3.32SSSNTVVSVGLNPADD482 pKa = 3.57ANLFRR487 pKa = 11.84WLSGIARR494 pKa = 11.84RR495 pKa = 11.84YY496 pKa = 7.87TMYY499 pKa = 10.1RR500 pKa = 11.84WKK502 pKa = 10.33KK503 pKa = 10.42LRR505 pKa = 11.84VLYY508 pKa = 10.61QSTCPTTTPGSVHH521 pKa = 6.94LGLFYY526 pKa = 10.92DD527 pKa = 5.85AEE529 pKa = 4.52DD530 pKa = 3.91LGNWIADD537 pKa = 3.53GSSTVTLSQTVGASIGPTWGSTMTSTNSGSITTNMVEE574 pKa = 3.96VDD576 pKa = 3.29VTRR579 pKa = 11.84AHH581 pKa = 6.5QRR583 pKa = 11.84VSWHH587 pKa = 6.81LIDD590 pKa = 5.25GNTGGTAVDD599 pKa = 3.94NQSVAALCASVVTPVPGGAVVGRR622 pKa = 11.84IWYY625 pKa = 9.3DD626 pKa = 3.3YY627 pKa = 10.34EE628 pKa = 5.14IEE630 pKa = 4.92LIQPTFAFGGNGLRR644 pKa = 11.84GGAGARR650 pKa = 11.84GFDD653 pKa = 3.79PSKK656 pKa = 10.77DD657 pKa = 3.26IYY659 pKa = 9.53LTKK662 pKa = 9.8PDD664 pKa = 4.25YY665 pKa = 11.41VPVPRR670 pKa = 11.84PLPKK674 pKa = 9.11PTPSGDD680 pKa = 3.37DD681 pKa = 3.46DD682 pKa = 4.17EE683 pKa = 5.3EE684 pKa = 5.39RR685 pKa = 11.84EE686 pKa = 4.29PVGG689 pKa = 4.09
MM1 pKa = 7.25EE2 pKa = 5.14QFICSYY8 pKa = 10.65KK9 pKa = 10.14GAKK12 pKa = 7.95QHH14 pKa = 5.24TVRR17 pKa = 11.84AAAEE21 pKa = 4.0DD22 pKa = 4.25VLRR25 pKa = 11.84HH26 pKa = 4.98PAVRR30 pKa = 11.84RR31 pKa = 11.84DD32 pKa = 3.53AKK34 pKa = 10.95LKK36 pKa = 9.82TFVKK40 pKa = 10.52AEE42 pKa = 3.94KK43 pKa = 10.48LNLSSKK49 pKa = 9.85PDD51 pKa = 3.65PAPRR55 pKa = 11.84VIQPRR60 pKa = 11.84DD61 pKa = 3.16VRR63 pKa = 11.84YY64 pKa = 8.53NCEE67 pKa = 3.54VGPYY71 pKa = 9.88LKK73 pKa = 10.6AHH75 pKa = 6.01EE76 pKa = 4.09HH77 pKa = 5.56TIYY80 pKa = 10.67RR81 pKa = 11.84GIAKK85 pKa = 9.48IWGGPTVMKK94 pKa = 10.53GFNSVDD100 pKa = 3.38TAKK103 pKa = 10.5HH104 pKa = 4.72MRR106 pKa = 11.84EE107 pKa = 4.02MWDD110 pKa = 4.38SIHH113 pKa = 7.11DD114 pKa = 3.73PCGIGLDD121 pKa = 3.45ASRR124 pKa = 11.84FDD126 pKa = 3.33QHH128 pKa = 7.84VSEE131 pKa = 4.65EE132 pKa = 4.04ALRR135 pKa = 11.84WEE137 pKa = 4.0HH138 pKa = 6.29SVYY141 pKa = 11.32NGMFKK146 pKa = 10.46SKK148 pKa = 10.46RR149 pKa = 11.84LAEE152 pKa = 4.12LLTWQINNVGTSYY165 pKa = 10.32TPEE168 pKa = 3.56GRR170 pKa = 11.84VTYY173 pKa = 9.83KK174 pKa = 10.97VKK176 pKa = 9.91GCRR179 pKa = 11.84MSGDD183 pKa = 3.68MNTSLGNCLIMCGMVWEE200 pKa = 5.2LCQQLGCPARR210 pKa = 11.84LANNGDD216 pKa = 3.82DD217 pKa = 4.47CMLIVPRR224 pKa = 11.84SWEE227 pKa = 3.43QRR229 pKa = 11.84VRR231 pKa = 11.84DD232 pKa = 4.7KK233 pKa = 10.83IASWFLDD240 pKa = 3.47FGFTMKK246 pKa = 10.55VEE248 pKa = 3.99DD249 pKa = 3.54TVYY252 pKa = 10.43EE253 pKa = 4.07FEE255 pKa = 5.08QIEE258 pKa = 4.63FCQTRR263 pKa = 11.84PVFASNRR270 pKa = 11.84WVMTRR275 pKa = 11.84DD276 pKa = 3.27YY277 pKa = 10.65RR278 pKa = 11.84VVMDD282 pKa = 4.99KK283 pKa = 10.58DD284 pKa = 4.19TFCLHH289 pKa = 7.44PDD291 pKa = 3.06NMPYY295 pKa = 10.22EE296 pKa = 3.97RR297 pKa = 11.84WLRR300 pKa = 11.84GVGTAGLALASGVPVLQTFYY320 pKa = 10.0TQLVALGGGQGEE332 pKa = 5.01LIDD335 pKa = 4.33GSGMSYY341 pKa = 8.8MAKK344 pKa = 10.39GLTPDD349 pKa = 4.06ASPITTEE356 pKa = 4.1ARR358 pKa = 11.84VSFYY362 pKa = 10.79KK363 pKa = 10.79AFGMPPWVQEE373 pKa = 4.03AYY375 pKa = 9.99EE376 pKa = 4.08EE377 pKa = 4.61EE378 pKa = 4.48IVASASSTNLGLIGSRR394 pKa = 11.84YY395 pKa = 7.53TYY397 pKa = 10.88HH398 pKa = 6.89NNTLAFQGMTGKK410 pKa = 8.88MKK412 pKa = 10.62KK413 pKa = 9.98PSTQKK418 pKa = 11.24SMIRR422 pKa = 11.84NIPIDD427 pKa = 3.92VPGGPPGYY435 pKa = 9.5TNFQRR440 pKa = 11.84KK441 pKa = 6.43TGPRR445 pKa = 11.84VTATKK450 pKa = 10.6AGLQIANSEE459 pKa = 4.08QFLPLSADD467 pKa = 3.32SSSNTVVSVGLNPADD482 pKa = 3.57ANLFRR487 pKa = 11.84WLSGIARR494 pKa = 11.84RR495 pKa = 11.84YY496 pKa = 7.87TMYY499 pKa = 10.1RR500 pKa = 11.84WKK502 pKa = 10.33KK503 pKa = 10.42LRR505 pKa = 11.84VLYY508 pKa = 10.61QSTCPTTTPGSVHH521 pKa = 6.94LGLFYY526 pKa = 10.92DD527 pKa = 5.85AEE529 pKa = 4.52DD530 pKa = 3.91LGNWIADD537 pKa = 3.53GSSTVTLSQTVGASIGPTWGSTMTSTNSGSITTNMVEE574 pKa = 3.96VDD576 pKa = 3.29VTRR579 pKa = 11.84AHH581 pKa = 6.5QRR583 pKa = 11.84VSWHH587 pKa = 6.81LIDD590 pKa = 5.25GNTGGTAVDD599 pKa = 3.94NQSVAALCASVVTPVPGGAVVGRR622 pKa = 11.84IWYY625 pKa = 9.3DD626 pKa = 3.3YY627 pKa = 10.34EE628 pKa = 5.14IEE630 pKa = 4.92LIQPTFAFGGNGLRR644 pKa = 11.84GGAGARR650 pKa = 11.84GFDD653 pKa = 3.79PSKK656 pKa = 10.77DD657 pKa = 3.26IYY659 pKa = 9.53LTKK662 pKa = 9.8PDD664 pKa = 4.25YY665 pKa = 11.41VPVPRR670 pKa = 11.84PLPKK674 pKa = 9.11PTPSGDD680 pKa = 3.37DD681 pKa = 3.46DD682 pKa = 4.17EE683 pKa = 5.3EE684 pKa = 5.39RR685 pKa = 11.84EE686 pKa = 4.29PVGG689 pKa = 4.09
Molecular weight: 76.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG67|A0A1L3KG67_9VIRU Uncharacterized protein OS=Shahe tombus-like virus 2 OX=1923456 PE=4 SV=1
MM1 pKa = 7.9ADD3 pKa = 4.28FSPVPAATIGGEE15 pKa = 4.25SQSEE19 pKa = 4.22GMANPNFTFGTVEE32 pKa = 3.8VLARR36 pKa = 11.84GAVACIVLTGRR47 pKa = 11.84KK48 pKa = 8.55LSKK51 pKa = 10.04WFWPYY56 pKa = 11.01QPTAKK61 pKa = 10.56QIDD64 pKa = 4.28GLVKK68 pKa = 10.41LDD70 pKa = 4.7SEE72 pKa = 4.94DD73 pKa = 3.52NDD75 pKa = 4.03GGVSGWALTKK85 pKa = 10.09KK86 pKa = 8.78WCAVAKK92 pKa = 10.77AEE94 pKa = 4.46FGTVQKK100 pKa = 9.5RR101 pKa = 11.84TPANDD106 pKa = 2.99AAIADD111 pKa = 4.32KK112 pKa = 10.53IRR114 pKa = 11.84RR115 pKa = 11.84LMKK118 pKa = 9.92EE119 pKa = 3.53ANTRR123 pKa = 11.84NVDD126 pKa = 4.13IIRR129 pKa = 11.84VLPWAVASVYY139 pKa = 10.66VPTDD143 pKa = 3.27ADD145 pKa = 3.63LVAASVYY152 pKa = 7.6NHH154 pKa = 6.5KK155 pKa = 10.31VVKK158 pKa = 10.32ARR160 pKa = 11.84RR161 pKa = 11.84KK162 pKa = 9.75HH163 pKa = 5.42LAVPQKK169 pKa = 11.06
MM1 pKa = 7.9ADD3 pKa = 4.28FSPVPAATIGGEE15 pKa = 4.25SQSEE19 pKa = 4.22GMANPNFTFGTVEE32 pKa = 3.8VLARR36 pKa = 11.84GAVACIVLTGRR47 pKa = 11.84KK48 pKa = 8.55LSKK51 pKa = 10.04WFWPYY56 pKa = 11.01QPTAKK61 pKa = 10.56QIDD64 pKa = 4.28GLVKK68 pKa = 10.41LDD70 pKa = 4.7SEE72 pKa = 4.94DD73 pKa = 3.52NDD75 pKa = 4.03GGVSGWALTKK85 pKa = 10.09KK86 pKa = 8.78WCAVAKK92 pKa = 10.77AEE94 pKa = 4.46FGTVQKK100 pKa = 9.5RR101 pKa = 11.84TPANDD106 pKa = 2.99AAIADD111 pKa = 4.32KK112 pKa = 10.53IRR114 pKa = 11.84RR115 pKa = 11.84LMKK118 pKa = 9.92EE119 pKa = 3.53ANTRR123 pKa = 11.84NVDD126 pKa = 4.13IIRR129 pKa = 11.84VLPWAVASVYY139 pKa = 10.66VPTDD143 pKa = 3.27ADD145 pKa = 3.63LVAASVYY152 pKa = 7.6NHH154 pKa = 6.5KK155 pKa = 10.31VVKK158 pKa = 10.32ARR160 pKa = 11.84RR161 pKa = 11.84KK162 pKa = 9.75HH163 pKa = 5.42LAVPQKK169 pKa = 11.06
Molecular weight: 18.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
858 |
169 |
689 |
429.0 |
47.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.508 ± 2.699 | 1.748 ± 0.268 |
5.478 ± 0.208 | 4.429 ± 0.417 |
3.263 ± 0.145 | 8.741 ± 1.058 |
1.748 ± 0.268 | 4.196 ± 0.026 |
5.245 ± 1.441 | 6.643 ± 0.344 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.03 ± 0.595 | 3.963 ± 0.085 |
5.944 ± 0.293 | 3.38 ± 0.2 |
5.828 ± 0.238 | 6.527 ± 0.85 |
7.226 ± 0.621 | 8.392 ± 1.352 |
2.448 ± 0.242 | 3.263 ± 0.706 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
