
Hericium alpestre
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Russulales; Hericiaceae; Hericium
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
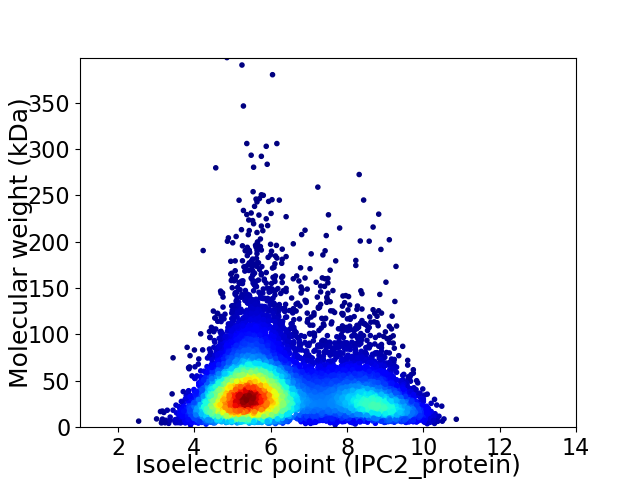
Virtual 2D-PAGE plot for 11000 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A4Z0AB03|A0A4Z0AB03_9AGAM CID domain-containing protein OS=Hericium alpestre OX=135208 GN=EWM64_g256 PE=4 SV=1
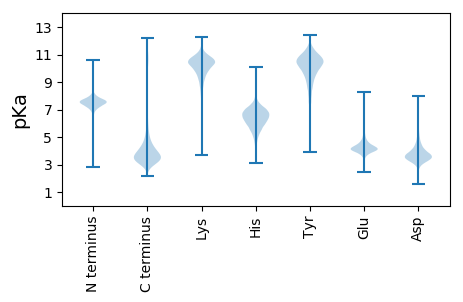
MM1 pKa = 7.58IFGIVYY7 pKa = 9.68DD8 pKa = 4.04AEE10 pKa = 4.3SVRR13 pKa = 11.84IVAHH17 pKa = 5.75VPYY20 pKa = 9.86EE21 pKa = 4.09IPDD24 pKa = 3.36EE25 pKa = 4.77DD26 pKa = 4.02GQAKK30 pKa = 9.24CQYY33 pKa = 9.46PRR35 pKa = 11.84YY36 pKa = 9.98SYY38 pKa = 9.22MSCVIDD44 pKa = 3.94TISYY48 pKa = 9.92RR49 pKa = 11.84GGRR52 pKa = 11.84RR53 pKa = 11.84CDD55 pKa = 3.04KK56 pKa = 10.7TSALDD61 pKa = 3.48RR62 pKa = 11.84FRR64 pKa = 11.84VAMAMVCLGIHH75 pKa = 6.55AEE77 pKa = 4.14FLAALLDD84 pKa = 4.62PSTSWPEE91 pKa = 3.96DD92 pKa = 3.29VVDD95 pKa = 3.64AAEE98 pKa = 4.35SVRR101 pKa = 11.84LLYY104 pKa = 10.59QPTLPTTSDD113 pKa = 3.39EE114 pKa = 4.3DD115 pKa = 4.05ASDD118 pKa = 4.61YY119 pKa = 11.53EE120 pKa = 4.65DD121 pKa = 4.14TPDD124 pKa = 5.47DD125 pKa = 3.74NTFDD129 pKa = 6.07DD130 pKa = 4.5EE131 pKa = 4.81DD132 pKa = 3.9TDD134 pKa = 3.82EE135 pKa = 6.1DD136 pKa = 4.0EE137 pKa = 4.95EE138 pKa = 4.48EE139 pKa = 4.39VAGEE143 pKa = 4.23CRR145 pKa = 3.56
MM1 pKa = 7.58IFGIVYY7 pKa = 9.68DD8 pKa = 4.04AEE10 pKa = 4.3SVRR13 pKa = 11.84IVAHH17 pKa = 5.75VPYY20 pKa = 9.86EE21 pKa = 4.09IPDD24 pKa = 3.36EE25 pKa = 4.77DD26 pKa = 4.02GQAKK30 pKa = 9.24CQYY33 pKa = 9.46PRR35 pKa = 11.84YY36 pKa = 9.98SYY38 pKa = 9.22MSCVIDD44 pKa = 3.94TISYY48 pKa = 9.92RR49 pKa = 11.84GGRR52 pKa = 11.84RR53 pKa = 11.84CDD55 pKa = 3.04KK56 pKa = 10.7TSALDD61 pKa = 3.48RR62 pKa = 11.84FRR64 pKa = 11.84VAMAMVCLGIHH75 pKa = 6.55AEE77 pKa = 4.14FLAALLDD84 pKa = 4.62PSTSWPEE91 pKa = 3.96DD92 pKa = 3.29VVDD95 pKa = 3.64AAEE98 pKa = 4.35SVRR101 pKa = 11.84LLYY104 pKa = 10.59QPTLPTTSDD113 pKa = 3.39EE114 pKa = 4.3DD115 pKa = 4.05ASDD118 pKa = 4.61YY119 pKa = 11.53EE120 pKa = 4.65DD121 pKa = 4.14TPDD124 pKa = 5.47DD125 pKa = 3.74NTFDD129 pKa = 6.07DD130 pKa = 4.5EE131 pKa = 4.81DD132 pKa = 3.9TDD134 pKa = 3.82EE135 pKa = 6.1DD136 pKa = 4.0EE137 pKa = 4.95EE138 pKa = 4.48EE139 pKa = 4.39VAGEE143 pKa = 4.23CRR145 pKa = 3.56
Molecular weight: 16.29 kDa
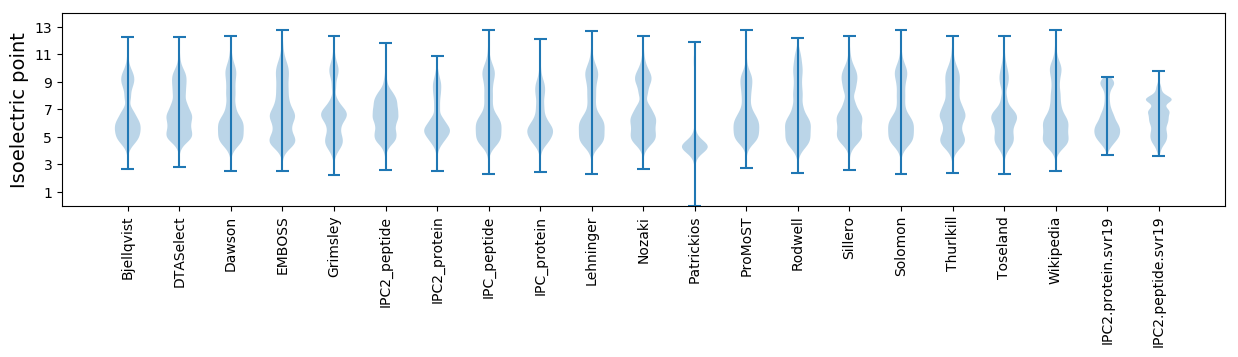
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y9ZLW2|A0A4Y9ZLW2_9AGAM Tr-type G domain-containing protein OS=Hericium alpestre OX=135208 GN=EWM64_g8924 PE=4 SV=1
MM1 pKa = 7.2TVTVPRR7 pKa = 11.84TRR9 pKa = 11.84MHH11 pKa = 6.73RR12 pKa = 11.84SKK14 pKa = 9.44TLSLAAPRR22 pKa = 11.84APSFTTLPTEE32 pKa = 4.97LILDD36 pKa = 3.6ILEE39 pKa = 4.49RR40 pKa = 11.84ALLASPKK47 pKa = 9.37TGHH50 pKa = 6.78AYY52 pKa = 10.48ARR54 pKa = 11.84INKK57 pKa = 9.11HH58 pKa = 4.98IAHH61 pKa = 7.41LFDD64 pKa = 4.09QILYY68 pKa = 8.07RR69 pKa = 11.84TVVLASSRR77 pKa = 11.84QIALFARR84 pKa = 11.84TLASKK89 pKa = 10.35PPSFLPAHH97 pKa = 6.3VKK99 pKa = 10.29RR100 pKa = 11.84LAITWDD106 pKa = 3.58TPTTLPATTHH116 pKa = 5.56AHH118 pKa = 5.66LTTIIHH124 pKa = 6.49ACAGTRR130 pKa = 11.84ALAVPQEE137 pKa = 4.04LMSICLEE144 pKa = 4.2ACPPAHH150 pKa = 6.8HH151 pKa = 7.22PAAHH155 pKa = 7.52DD156 pKa = 3.89YY157 pKa = 7.39PAPMPADD164 pKa = 3.23ITIGAFTDD172 pKa = 3.66MAPLFPLRR180 pKa = 11.84TEE182 pKa = 4.12KK183 pKa = 10.49QALTSSTSAPPLFAGVTRR201 pKa = 11.84LRR203 pKa = 11.84IAEE206 pKa = 4.13PAGTWTAPRR215 pKa = 11.84AMLAQLGPLAQLTHH229 pKa = 6.26LHH231 pKa = 6.83LARR234 pKa = 11.84ARR236 pKa = 11.84ARR238 pKa = 11.84MRR240 pKa = 11.84TTTSRR245 pKa = 11.84SRR247 pKa = 11.84RR248 pKa = 11.84TSPDD252 pKa = 2.83CSGPGAGVDD261 pKa = 3.59PVKK264 pKa = 10.63EE265 pKa = 3.94IGSAEE270 pKa = 4.07LNHH273 pKa = 6.65
MM1 pKa = 7.2TVTVPRR7 pKa = 11.84TRR9 pKa = 11.84MHH11 pKa = 6.73RR12 pKa = 11.84SKK14 pKa = 9.44TLSLAAPRR22 pKa = 11.84APSFTTLPTEE32 pKa = 4.97LILDD36 pKa = 3.6ILEE39 pKa = 4.49RR40 pKa = 11.84ALLASPKK47 pKa = 9.37TGHH50 pKa = 6.78AYY52 pKa = 10.48ARR54 pKa = 11.84INKK57 pKa = 9.11HH58 pKa = 4.98IAHH61 pKa = 7.41LFDD64 pKa = 4.09QILYY68 pKa = 8.07RR69 pKa = 11.84TVVLASSRR77 pKa = 11.84QIALFARR84 pKa = 11.84TLASKK89 pKa = 10.35PPSFLPAHH97 pKa = 6.3VKK99 pKa = 10.29RR100 pKa = 11.84LAITWDD106 pKa = 3.58TPTTLPATTHH116 pKa = 5.56AHH118 pKa = 5.66LTTIIHH124 pKa = 6.49ACAGTRR130 pKa = 11.84ALAVPQEE137 pKa = 4.04LMSICLEE144 pKa = 4.2ACPPAHH150 pKa = 6.8HH151 pKa = 7.22PAAHH155 pKa = 7.52DD156 pKa = 3.89YY157 pKa = 7.39PAPMPADD164 pKa = 3.23ITIGAFTDD172 pKa = 3.66MAPLFPLRR180 pKa = 11.84TEE182 pKa = 4.12KK183 pKa = 10.49QALTSSTSAPPLFAGVTRR201 pKa = 11.84LRR203 pKa = 11.84IAEE206 pKa = 4.13PAGTWTAPRR215 pKa = 11.84AMLAQLGPLAQLTHH229 pKa = 6.26LHH231 pKa = 6.83LARR234 pKa = 11.84ARR236 pKa = 11.84ARR238 pKa = 11.84MRR240 pKa = 11.84TTTSRR245 pKa = 11.84SRR247 pKa = 11.84RR248 pKa = 11.84TSPDD252 pKa = 2.83CSGPGAGVDD261 pKa = 3.59PVKK264 pKa = 10.63EE265 pKa = 3.94IGSAEE270 pKa = 4.07LNHH273 pKa = 6.65
Molecular weight: 29.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4291909 |
20 |
3630 |
390.2 |
43.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.24 ± 0.022 | 1.196 ± 0.009 |
5.97 ± 0.017 | 6.19 ± 0.022 |
3.716 ± 0.014 | 6.385 ± 0.02 |
2.626 ± 0.01 | 4.755 ± 0.018 |
4.679 ± 0.026 | 9.189 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.213 ± 0.01 | 3.224 ± 0.013 |
6.41 ± 0.033 | 3.755 ± 0.014 |
6.092 ± 0.021 | 7.989 ± 0.031 |
5.743 ± 0.016 | 6.429 ± 0.018 |
1.437 ± 0.01 | 2.609 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
