
Methylobacterium sp. Leaf93
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Methylobacterium; unclassified Methylobacterium
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
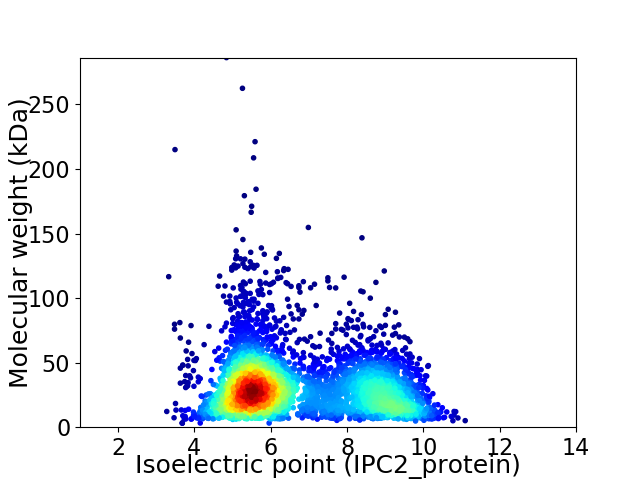
Virtual 2D-PAGE plot for 4228 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q4XF70|A0A0Q4XF70_9RHIZ NAD-dependent dehydratase OS=Methylobacterium sp. Leaf93 OX=1736249 GN=ASF26_09580 PE=4 SV=1
MM1 pKa = 7.22NWIEE5 pKa = 4.22LPKK8 pKa = 10.74LPTYY12 pKa = 9.69WDD14 pKa = 4.4EE15 pKa = 4.32IDD17 pKa = 4.16GSEE20 pKa = 3.82SHH22 pKa = 6.17YY23 pKa = 11.0QFGYY27 pKa = 11.09DD28 pKa = 4.99DD29 pKa = 4.92GDD31 pKa = 3.91SGDD34 pKa = 3.63SMLYY38 pKa = 9.9SGSNFKK44 pKa = 10.93YY45 pKa = 10.18YY46 pKa = 10.62EE47 pKa = 4.29EE48 pKa = 4.45YY49 pKa = 10.81DD50 pKa = 3.24EE51 pKa = 5.3GEE53 pKa = 3.79IYY55 pKa = 10.62VYY57 pKa = 10.71YY58 pKa = 10.59DD59 pKa = 3.17QIISGNIKK67 pKa = 9.65SVNYY71 pKa = 10.31NYY73 pKa = 10.44YY74 pKa = 9.94QYY76 pKa = 11.13EE77 pKa = 3.95EE78 pKa = 4.77EE79 pKa = 4.49YY80 pKa = 11.33YY81 pKa = 10.47EE82 pKa = 6.0DD83 pKa = 3.59EE84 pKa = 5.0DD85 pKa = 4.91GYY87 pKa = 11.39EE88 pKa = 3.79FSIDD92 pKa = 3.3VRR94 pKa = 11.84YY95 pKa = 10.28EE96 pKa = 3.84GAFEE100 pKa = 4.0LTKK103 pKa = 10.22IDD105 pKa = 3.39VDD107 pKa = 4.16FSEE110 pKa = 4.08ILGRR114 pKa = 11.84SAEE117 pKa = 4.23DD118 pKa = 3.16NRR120 pKa = 11.84QLVFRR125 pKa = 11.84GNDD128 pKa = 3.32QLIGSAANDD137 pKa = 3.57SLIGYY142 pKa = 8.69DD143 pKa = 4.23GNDD146 pKa = 3.06RR147 pKa = 11.84LYY149 pKa = 11.29GGGGDD154 pKa = 4.8DD155 pKa = 5.67SLDD158 pKa = 3.76GGNGSDD164 pKa = 5.06SDD166 pKa = 3.99YY167 pKa = 11.38LGNDD171 pKa = 3.12KK172 pKa = 11.12LYY174 pKa = 11.1GGSGNDD180 pKa = 3.55KK181 pKa = 10.57LAGRR185 pKa = 11.84YY186 pKa = 9.74GDD188 pKa = 4.67DD189 pKa = 3.65RR190 pKa = 11.84LEE192 pKa = 4.46GGNGNDD198 pKa = 4.44KK199 pKa = 11.03LDD201 pKa = 4.1AGSGNDD207 pKa = 3.59TLLGQDD213 pKa = 3.97GDD215 pKa = 4.83DD216 pKa = 4.03NLTSGKK222 pKa = 8.68GTDD225 pKa = 3.76ALLGGAGSDD234 pKa = 3.52ILNGGADD241 pKa = 4.12KK242 pKa = 10.01DD243 pKa = 4.18TLSGGGGKK251 pKa = 10.48GLDD254 pKa = 3.28TFVFALDD261 pKa = 3.59KK262 pKa = 10.3TAGAISRR269 pKa = 11.84SDD271 pKa = 4.47TILDD275 pKa = 3.46WNPTYY280 pKa = 11.16DD281 pKa = 5.28SIDD284 pKa = 3.35LKK286 pKa = 11.39YY287 pKa = 10.92AGTRR291 pKa = 11.84DD292 pKa = 3.31NYY294 pKa = 10.72FEE296 pKa = 4.39TRR298 pKa = 11.84TTTISAEE305 pKa = 3.95MAYY308 pKa = 10.47ARR310 pKa = 11.84YY311 pKa = 10.31DD312 pKa = 3.58GVLKK316 pKa = 10.8NDD318 pKa = 3.9DD319 pKa = 3.67MTHH322 pKa = 6.45LFLYY326 pKa = 7.94NTSTKK331 pKa = 9.77TGYY334 pKa = 10.7LLSDD338 pKa = 3.45QNADD342 pKa = 3.18HH343 pKa = 6.64MFDD346 pKa = 3.44TVVVISNAASAASMSYY362 pKa = 11.13LDD364 pKa = 3.87IVV366 pKa = 3.58
MM1 pKa = 7.22NWIEE5 pKa = 4.22LPKK8 pKa = 10.74LPTYY12 pKa = 9.69WDD14 pKa = 4.4EE15 pKa = 4.32IDD17 pKa = 4.16GSEE20 pKa = 3.82SHH22 pKa = 6.17YY23 pKa = 11.0QFGYY27 pKa = 11.09DD28 pKa = 4.99DD29 pKa = 4.92GDD31 pKa = 3.91SGDD34 pKa = 3.63SMLYY38 pKa = 9.9SGSNFKK44 pKa = 10.93YY45 pKa = 10.18YY46 pKa = 10.62EE47 pKa = 4.29EE48 pKa = 4.45YY49 pKa = 10.81DD50 pKa = 3.24EE51 pKa = 5.3GEE53 pKa = 3.79IYY55 pKa = 10.62VYY57 pKa = 10.71YY58 pKa = 10.59DD59 pKa = 3.17QIISGNIKK67 pKa = 9.65SVNYY71 pKa = 10.31NYY73 pKa = 10.44YY74 pKa = 9.94QYY76 pKa = 11.13EE77 pKa = 3.95EE78 pKa = 4.77EE79 pKa = 4.49YY80 pKa = 11.33YY81 pKa = 10.47EE82 pKa = 6.0DD83 pKa = 3.59EE84 pKa = 5.0DD85 pKa = 4.91GYY87 pKa = 11.39EE88 pKa = 3.79FSIDD92 pKa = 3.3VRR94 pKa = 11.84YY95 pKa = 10.28EE96 pKa = 3.84GAFEE100 pKa = 4.0LTKK103 pKa = 10.22IDD105 pKa = 3.39VDD107 pKa = 4.16FSEE110 pKa = 4.08ILGRR114 pKa = 11.84SAEE117 pKa = 4.23DD118 pKa = 3.16NRR120 pKa = 11.84QLVFRR125 pKa = 11.84GNDD128 pKa = 3.32QLIGSAANDD137 pKa = 3.57SLIGYY142 pKa = 8.69DD143 pKa = 4.23GNDD146 pKa = 3.06RR147 pKa = 11.84LYY149 pKa = 11.29GGGGDD154 pKa = 4.8DD155 pKa = 5.67SLDD158 pKa = 3.76GGNGSDD164 pKa = 5.06SDD166 pKa = 3.99YY167 pKa = 11.38LGNDD171 pKa = 3.12KK172 pKa = 11.12LYY174 pKa = 11.1GGSGNDD180 pKa = 3.55KK181 pKa = 10.57LAGRR185 pKa = 11.84YY186 pKa = 9.74GDD188 pKa = 4.67DD189 pKa = 3.65RR190 pKa = 11.84LEE192 pKa = 4.46GGNGNDD198 pKa = 4.44KK199 pKa = 11.03LDD201 pKa = 4.1AGSGNDD207 pKa = 3.59TLLGQDD213 pKa = 3.97GDD215 pKa = 4.83DD216 pKa = 4.03NLTSGKK222 pKa = 8.68GTDD225 pKa = 3.76ALLGGAGSDD234 pKa = 3.52ILNGGADD241 pKa = 4.12KK242 pKa = 10.01DD243 pKa = 4.18TLSGGGGKK251 pKa = 10.48GLDD254 pKa = 3.28TFVFALDD261 pKa = 3.59KK262 pKa = 10.3TAGAISRR269 pKa = 11.84SDD271 pKa = 4.47TILDD275 pKa = 3.46WNPTYY280 pKa = 11.16DD281 pKa = 5.28SIDD284 pKa = 3.35LKK286 pKa = 11.39YY287 pKa = 10.92AGTRR291 pKa = 11.84DD292 pKa = 3.31NYY294 pKa = 10.72FEE296 pKa = 4.39TRR298 pKa = 11.84TTTISAEE305 pKa = 3.95MAYY308 pKa = 10.47ARR310 pKa = 11.84YY311 pKa = 10.31DD312 pKa = 3.58GVLKK316 pKa = 10.8NDD318 pKa = 3.9DD319 pKa = 3.67MTHH322 pKa = 6.45LFLYY326 pKa = 7.94NTSTKK331 pKa = 9.77TGYY334 pKa = 10.7LLSDD338 pKa = 3.45QNADD342 pKa = 3.18HH343 pKa = 6.64MFDD346 pKa = 3.44TVVVISNAASAASMSYY362 pKa = 11.13LDD364 pKa = 3.87IVV366 pKa = 3.58
Molecular weight: 40.04 kDa
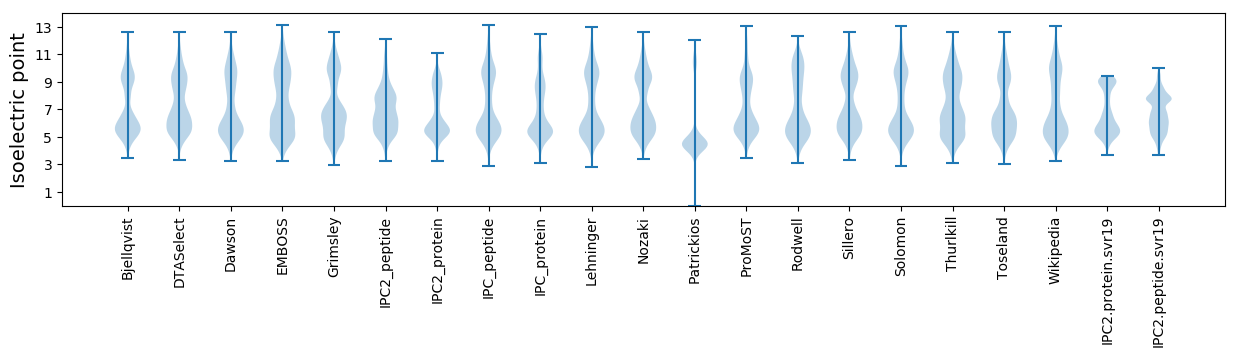
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q4X7V5|A0A0Q4X7V5_9RHIZ Response regulator receiver protein OS=Methylobacterium sp. Leaf93 OX=1736249 GN=ASF26_06430 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATAGGRR28 pKa = 11.84KK29 pKa = 9.07VIAARR34 pKa = 11.84RR35 pKa = 11.84SHH37 pKa = 5.5GRR39 pKa = 11.84KK40 pKa = 9.35KK41 pKa = 10.69LSAA44 pKa = 3.99
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATAGGRR28 pKa = 11.84KK29 pKa = 9.07VIAARR34 pKa = 11.84RR35 pKa = 11.84SHH37 pKa = 5.5GRR39 pKa = 11.84KK40 pKa = 9.35KK41 pKa = 10.69LSAA44 pKa = 3.99
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1315157 |
29 |
2686 |
311.1 |
33.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.356 ± 0.06 | 0.778 ± 0.013 |
5.791 ± 0.034 | 5.513 ± 0.039 |
3.456 ± 0.022 | 9.118 ± 0.044 |
1.939 ± 0.019 | 4.853 ± 0.024 |
2.769 ± 0.032 | 10.25 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.161 ± 0.016 | 2.234 ± 0.023 |
5.559 ± 0.032 | 2.701 ± 0.022 |
7.778 ± 0.043 | 5.418 ± 0.027 |
5.56 ± 0.032 | 7.537 ± 0.029 |
1.199 ± 0.015 | 2.03 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
