
Halomicrobium zhouii
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Haloarculaceae; Halomicrobium
Average proteome isoelectric point is 4.85
Get precalculated fractions of proteins
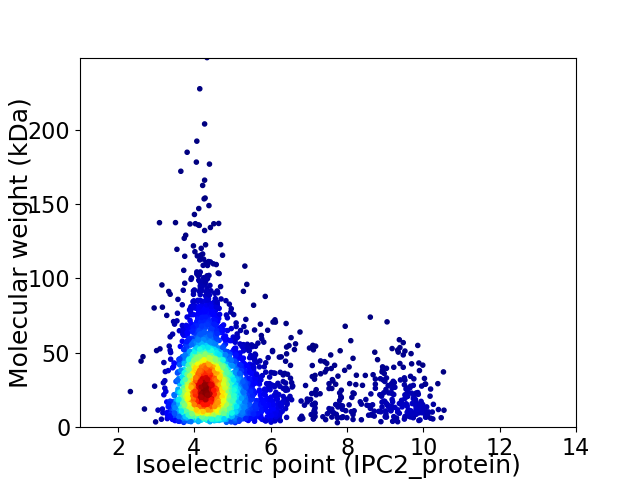
Virtual 2D-PAGE plot for 4091 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
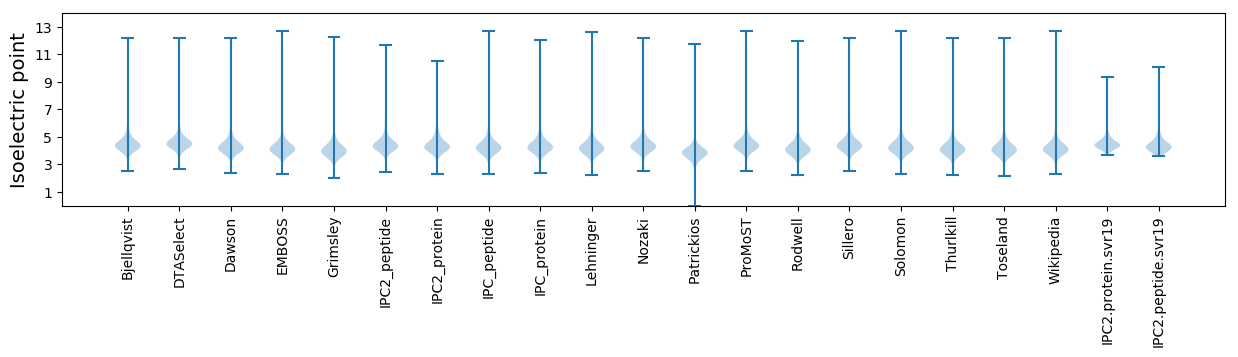
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I6K6R6|A0A1I6K6R6_9EURY Universal stress protein family protein OS=Halomicrobium zhouii OX=767519 GN=SAMN05216559_0234 PE=4 SV=1
MM1 pKa = 7.24TGVAEE6 pKa = 4.33VVDD9 pKa = 4.79GVTTTLLCGASLGGEE24 pKa = 3.79TRR26 pKa = 11.84EE27 pKa = 4.17CCEE30 pKa = 3.63ALLRR34 pKa = 11.84AGADD38 pKa = 3.56DD39 pKa = 3.73GRR41 pKa = 11.84EE42 pKa = 4.19VLWVTYY48 pKa = 7.29TRR50 pKa = 11.84PPADD54 pKa = 4.1CLASVPDD61 pKa = 4.03DD62 pKa = 3.78VSVRR66 pKa = 11.84GVLAVGDD73 pKa = 3.98APHH76 pKa = 7.37WEE78 pKa = 4.08TSIDD82 pKa = 3.82GVDD85 pKa = 3.68VEE87 pKa = 5.48VVATPGDD94 pKa = 3.54ITALGIKK101 pKa = 10.36LSRR104 pKa = 11.84FLSGGDD110 pKa = 4.14DD111 pKa = 3.99DD112 pKa = 5.39LTVCFDD118 pKa = 4.16SVTAMCQYY126 pKa = 10.62VEE128 pKa = 4.66PEE130 pKa = 3.69TAYY133 pKa = 10.75GFLHH137 pKa = 6.75TIAVQLYY144 pKa = 8.5AADD147 pKa = 3.79ATAHH151 pKa = 5.29FHH153 pKa = 6.84IDD155 pKa = 3.48PAAHH159 pKa = 7.02DD160 pKa = 4.31ASTVDD165 pKa = 5.43LFASLCDD172 pKa = 3.34AVVNVGDD179 pKa = 5.77DD180 pKa = 3.9GDD182 pKa = 4.87DD183 pKa = 3.88DD184 pKa = 4.28PAVRR188 pKa = 11.84TRR190 pKa = 11.84PVLDD194 pKa = 3.47
MM1 pKa = 7.24TGVAEE6 pKa = 4.33VVDD9 pKa = 4.79GVTTTLLCGASLGGEE24 pKa = 3.79TRR26 pKa = 11.84EE27 pKa = 4.17CCEE30 pKa = 3.63ALLRR34 pKa = 11.84AGADD38 pKa = 3.56DD39 pKa = 3.73GRR41 pKa = 11.84EE42 pKa = 4.19VLWVTYY48 pKa = 7.29TRR50 pKa = 11.84PPADD54 pKa = 4.1CLASVPDD61 pKa = 4.03DD62 pKa = 3.78VSVRR66 pKa = 11.84GVLAVGDD73 pKa = 3.98APHH76 pKa = 7.37WEE78 pKa = 4.08TSIDD82 pKa = 3.82GVDD85 pKa = 3.68VEE87 pKa = 5.48VVATPGDD94 pKa = 3.54ITALGIKK101 pKa = 10.36LSRR104 pKa = 11.84FLSGGDD110 pKa = 4.14DD111 pKa = 3.99DD112 pKa = 5.39LTVCFDD118 pKa = 4.16SVTAMCQYY126 pKa = 10.62VEE128 pKa = 4.66PEE130 pKa = 3.69TAYY133 pKa = 10.75GFLHH137 pKa = 6.75TIAVQLYY144 pKa = 8.5AADD147 pKa = 3.79ATAHH151 pKa = 5.29FHH153 pKa = 6.84IDD155 pKa = 3.48PAAHH159 pKa = 7.02DD160 pKa = 4.31ASTVDD165 pKa = 5.43LFASLCDD172 pKa = 3.34AVVNVGDD179 pKa = 5.77DD180 pKa = 3.9GDD182 pKa = 4.87DD183 pKa = 3.88DD184 pKa = 4.28PAVRR188 pKa = 11.84TRR190 pKa = 11.84PVLDD194 pKa = 3.47
Molecular weight: 20.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I6KZH9|A0A1I6KZH9_9EURY Uncharacterized protein OS=Halomicrobium zhouii OX=767519 GN=SAMN05216559_1664 PE=4 SV=1
MM1 pKa = 7.63LFPVRR6 pKa = 11.84FVVSPEE12 pKa = 3.34GSAAVLVAILAGVALGTASGLTPGLHH38 pKa = 6.86ANTFALGLAAVASSVPGPPTLVGATMLAAGTTHH71 pKa = 6.94TFLDD75 pKa = 4.09VVPALALGVPDD86 pKa = 4.19PAMAASALPGHH97 pKa = 6.7RR98 pKa = 11.84LVIAGRR104 pKa = 11.84GRR106 pKa = 11.84EE107 pKa = 4.26AIRR110 pKa = 11.84LSALGSGLAVVFAVPLAIPVTLAMQRR136 pKa = 11.84AYY138 pKa = 9.07PTIRR142 pKa = 11.84AHH144 pKa = 7.67LSLLLAGVAVLLVVTEE160 pKa = 4.37RR161 pKa = 11.84TPSGKK166 pKa = 10.32VGATLTLVASAALGFVTLDD185 pKa = 3.72LTPQAPLPVGGMLAPLFAGLFGAPVLIEE213 pKa = 4.04SLGGGGVPPQDD224 pKa = 3.95DD225 pKa = 3.51ATLAFSRR232 pKa = 11.84RR233 pKa = 11.84DD234 pKa = 3.48VGALALLGTGAGAIVGYY251 pKa = 10.2IPGVSAAIASAAALGVLSGSGPRR274 pKa = 11.84AFVVTTSGVNTATAIFALFALVALGSPRR302 pKa = 11.84TGVLVAVEE310 pKa = 4.22SAGIPIDD317 pKa = 4.35LPVLLASVGFAAVVAAVLVPVLGDD341 pKa = 4.53RR342 pKa = 11.84YY343 pKa = 10.57LDD345 pKa = 3.18WARR348 pKa = 11.84RR349 pKa = 11.84ANQNRR354 pKa = 11.84LAIGILSLLVALSFLFAGFVGIGAFGVATVVGSIPARR391 pKa = 11.84FGAKK395 pKa = 9.42RR396 pKa = 11.84ANCMGVLLGPLALSS410 pKa = 3.79
MM1 pKa = 7.63LFPVRR6 pKa = 11.84FVVSPEE12 pKa = 3.34GSAAVLVAILAGVALGTASGLTPGLHH38 pKa = 6.86ANTFALGLAAVASSVPGPPTLVGATMLAAGTTHH71 pKa = 6.94TFLDD75 pKa = 4.09VVPALALGVPDD86 pKa = 4.19PAMAASALPGHH97 pKa = 6.7RR98 pKa = 11.84LVIAGRR104 pKa = 11.84GRR106 pKa = 11.84EE107 pKa = 4.26AIRR110 pKa = 11.84LSALGSGLAVVFAVPLAIPVTLAMQRR136 pKa = 11.84AYY138 pKa = 9.07PTIRR142 pKa = 11.84AHH144 pKa = 7.67LSLLLAGVAVLLVVTEE160 pKa = 4.37RR161 pKa = 11.84TPSGKK166 pKa = 10.32VGATLTLVASAALGFVTLDD185 pKa = 3.72LTPQAPLPVGGMLAPLFAGLFGAPVLIEE213 pKa = 4.04SLGGGGVPPQDD224 pKa = 3.95DD225 pKa = 3.51ATLAFSRR232 pKa = 11.84RR233 pKa = 11.84DD234 pKa = 3.48VGALALLGTGAGAIVGYY251 pKa = 10.2IPGVSAAIASAAALGVLSGSGPRR274 pKa = 11.84AFVVTTSGVNTATAIFALFALVALGSPRR302 pKa = 11.84TGVLVAVEE310 pKa = 4.22SAGIPIDD317 pKa = 4.35LPVLLASVGFAAVVAAVLVPVLGDD341 pKa = 4.53RR342 pKa = 11.84YY343 pKa = 10.57LDD345 pKa = 3.18WARR348 pKa = 11.84RR349 pKa = 11.84ANQNRR354 pKa = 11.84LAIGILSLLVALSFLFAGFVGIGAFGVATVVGSIPARR391 pKa = 11.84FGAKK395 pKa = 9.42RR396 pKa = 11.84ANCMGVLLGPLALSS410 pKa = 3.79
Molecular weight: 40.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1202276 |
30 |
2225 |
293.9 |
31.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.904 ± 0.047 | 0.684 ± 0.013 |
8.823 ± 0.052 | 8.313 ± 0.054 |
3.335 ± 0.025 | 8.701 ± 0.041 |
2.001 ± 0.019 | 3.599 ± 0.028 |
1.649 ± 0.023 | 8.756 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.703 ± 0.015 | 2.315 ± 0.025 |
4.637 ± 0.028 | 2.668 ± 0.023 |
6.426 ± 0.034 | 5.612 ± 0.032 |
6.446 ± 0.041 | 9.518 ± 0.044 |
1.227 ± 0.016 | 2.683 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
