
Pseudomonas taeanensis MS-3
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas; Pseudomonas taeanensis
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
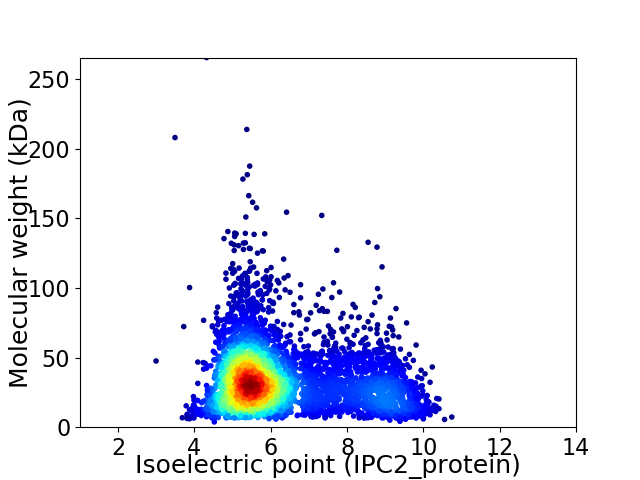
Virtual 2D-PAGE plot for 4573 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A1YFP6|A0A0A1YFP6_9PSED DeoR faimly transcriptional regulator OS=Pseudomonas taeanensis MS-3 OX=1395571 GN=TMS3_0114685 PE=4 SV=1
MM1 pKa = 7.46LLDD4 pKa = 3.74EE5 pKa = 4.69TAGALDD11 pKa = 3.83PTIGFPTAGLSFSPEE26 pKa = 3.84FPDD29 pKa = 5.17PEE31 pKa = 4.69LEE33 pKa = 4.26PDD35 pKa = 3.62VTPEE39 pKa = 3.43PTAAVVPEE47 pKa = 5.09GPDD50 pKa = 3.15STPEE54 pKa = 3.7VEE56 pKa = 5.96IIHH59 pKa = 6.19QDD61 pKa = 3.01AEE63 pKa = 4.56GQVLGAPGVVDD74 pKa = 4.43EE75 pKa = 5.18AALATGSNAASNAEE89 pKa = 3.98QAFGRR94 pKa = 11.84LVVNSPDD101 pKa = 3.17GVAALQVQDD110 pKa = 5.3KK111 pKa = 10.43DD112 pKa = 4.33GNWVDD117 pKa = 3.62VTAGGTVQGVYY128 pKa = 10.92GSLVVDD134 pKa = 4.1AAGRR138 pKa = 11.84WVYY141 pKa = 9.37TLNGSTQDD149 pKa = 3.49HH150 pKa = 6.84GNPSAIGAADD160 pKa = 3.41QVGEE164 pKa = 4.24SFAVRR169 pKa = 11.84VFDD172 pKa = 3.59TDD174 pKa = 3.79GDD176 pKa = 4.01VSPVSTLSININDD189 pKa = 4.78DD190 pKa = 3.39GPNAVDD196 pKa = 4.21DD197 pKa = 4.56SNSIAEE203 pKa = 4.36DD204 pKa = 3.45TLTPISGNVLSNDD217 pKa = 3.44LHH219 pKa = 7.1PNGQPGADD227 pKa = 3.43TPISFVSWTSTAATYY242 pKa = 7.93GTFTDD247 pKa = 4.4TGNGTYY253 pKa = 9.97SYY255 pKa = 11.06SLNNANPAVQGLDD268 pKa = 3.1AGEE271 pKa = 4.39SLSEE275 pKa = 4.17TFSYY279 pKa = 10.45TMQDD283 pKa = 2.85ADD285 pKa = 4.37GDD287 pKa = 4.28TATATLTITITGSNDD302 pKa = 3.14VPVLTVDD309 pKa = 4.82PGNQGANDD317 pKa = 3.64RR318 pKa = 11.84VFEE321 pKa = 4.73AGLAQGSDD329 pKa = 3.03AASNSEE335 pKa = 4.07FASGTFTLSDD345 pKa = 3.83ADD347 pKa = 4.62GLDD350 pKa = 3.99DD351 pKa = 4.69LQSVTINGVTVAIGSLAGSQFIGAHH376 pKa = 5.67GSLTISAYY384 pKa = 9.04NTATGVASYY393 pKa = 10.35SYY395 pKa = 10.54EE396 pKa = 4.05LTSPTTDD403 pKa = 3.14GPGIEE408 pKa = 4.45SDD410 pKa = 3.76SFTLSVSDD418 pKa = 3.92GTASSAPASIVIEE431 pKa = 4.05IVDD434 pKa = 4.32DD435 pKa = 4.02VPHH438 pKa = 7.02AVDD441 pKa = 4.76DD442 pKa = 4.54SNATFASEE450 pKa = 4.08NQLTLIGNVLDD461 pKa = 4.73NDD463 pKa = 3.86VQGADD468 pKa = 4.2RR469 pKa = 11.84LPGGPIQPAVMQGTYY484 pKa = 8.95GTLTLNADD492 pKa = 3.47GSYY495 pKa = 10.73SYY497 pKa = 11.45LLDD500 pKa = 4.4PSDD503 pKa = 4.96ADD505 pKa = 3.97FLALGGGGMATEE517 pKa = 4.72TFTYY521 pKa = 9.52TLNDD525 pKa = 3.66ADD527 pKa = 4.77GDD529 pKa = 4.2SDD531 pKa = 4.0TANLVLKK538 pKa = 10.34VRR540 pKa = 11.84NLDD543 pKa = 3.59DD544 pKa = 3.6KK545 pKa = 11.81VQIKK549 pKa = 10.69DD550 pKa = 3.19LDD552 pKa = 3.81VRR554 pKa = 11.84GGEE557 pKa = 4.09HH558 pKa = 7.05RR559 pKa = 11.84VDD561 pKa = 3.86EE562 pKa = 5.62DD563 pKa = 3.93DD564 pKa = 4.84LAMGSDD570 pKa = 3.81PSKK573 pKa = 10.99EE574 pKa = 4.06STTVSGTFKK583 pKa = 10.86VQAPDD588 pKa = 3.17GLLNLTVGGIQVVLNGVVSGFPQSVTTPLGNQLTITGFDD627 pKa = 3.67ADD629 pKa = 5.73DD630 pKa = 5.42GILDD634 pKa = 3.99EE635 pKa = 5.97GIVSYY640 pKa = 10.82SYY642 pKa = 10.23TLLGSAQHH650 pKa = 6.46APGGGEE656 pKa = 3.91NQLTEE661 pKa = 4.26HH662 pKa = 6.43FAVLAEE668 pKa = 4.71DD669 pKa = 3.94VDD671 pKa = 4.15HH672 pKa = 7.23DD673 pKa = 4.26TDD675 pKa = 5.38LASLDD680 pKa = 3.55INIIDD685 pKa = 4.48DD686 pKa = 3.74VPHH689 pKa = 6.91AVDD692 pKa = 4.43DD693 pKa = 4.54SNSIAEE699 pKa = 4.47DD700 pKa = 3.42SVTPITGNVLSNDD713 pKa = 3.59LHH715 pKa = 7.1PNGQPGADD723 pKa = 3.37TPTSFVGWTSTAATYY738 pKa = 7.93GTFTDD743 pKa = 4.4TGNGTYY749 pKa = 9.97SYY751 pKa = 11.06SLNNANPAVQGLNTGEE767 pKa = 4.3SLSEE771 pKa = 3.96TFTYY775 pKa = 9.93TMRR778 pKa = 11.84DD779 pKa = 3.15ADD781 pKa = 4.37GDD783 pKa = 3.96TATAKK788 pKa = 9.58LTITINGADD797 pKa = 3.59DD798 pKa = 3.67HH799 pKa = 7.44ASVVTAAAYY808 pKa = 10.16GPDD811 pKa = 3.07ATVYY815 pKa = 9.82EE816 pKa = 4.62HH817 pKa = 7.16GLTSLADD824 pKa = 3.75TSEE827 pKa = 4.3STSGSFTVSASDD839 pKa = 4.65GIASVTVGGATFSLAQLQALAATNQLVSTGEE870 pKa = 4.06GTLTLTGYY878 pKa = 10.07SGSATAGTLSYY889 pKa = 10.89SYY891 pKa = 9.71TLNATIDD898 pKa = 3.87NDD900 pKa = 3.68SHH902 pKa = 8.18AGATDD907 pKa = 3.07THH909 pKa = 7.4FDD911 pKa = 3.68DD912 pKa = 4.41SLSISVLGVGGSTASDD928 pKa = 3.4EE929 pKa = 4.27LVVRR933 pKa = 11.84IIDD936 pKa = 3.88DD937 pKa = 3.8TPTAMNDD944 pKa = 3.43GPVGVTEE951 pKa = 4.44DD952 pKa = 3.62AVSNSVGGNVLTNDD966 pKa = 3.35SAGADD971 pKa = 3.61TPKK974 pKa = 11.09AFTTWAATGHH984 pKa = 7.0DD985 pKa = 3.49NTAAINALNSFGTLTLAAGGVYY1007 pKa = 10.41SFVLDD1012 pKa = 3.54NSLASVQALNASSHH1026 pKa = 5.97LSYY1029 pKa = 11.04DD1030 pKa = 3.22VYY1032 pKa = 11.01YY1033 pKa = 10.86TMQDD1037 pKa = 2.88ADD1039 pKa = 4.36GDD1041 pKa = 4.2SSSAKK1046 pKa = 9.41LTITISGADD1055 pKa = 3.43DD1056 pKa = 3.56HH1057 pKa = 7.52ASVVTAAAYY1066 pKa = 10.16GPDD1069 pKa = 3.07ATVYY1073 pKa = 9.82EE1074 pKa = 4.62HH1075 pKa = 7.16GLTSLADD1082 pKa = 3.68TSEE1085 pKa = 4.31TTSGSFTVSASDD1097 pKa = 4.65GIASVTVGGATFSFAQLQALATTNQQVSTGEE1128 pKa = 4.14GTLTLSNYY1136 pKa = 10.1SGSATAGTLSYY1147 pKa = 10.89SYY1149 pKa = 9.71TLNATIDD1156 pKa = 3.72NDD1158 pKa = 3.9SHH1160 pKa = 8.04VGATGTYY1167 pKa = 9.76FDD1169 pKa = 5.49DD1170 pKa = 3.94SLSISVMGVGGSAASDD1186 pKa = 3.59DD1187 pKa = 4.01LVVRR1191 pKa = 11.84IVDD1194 pKa = 3.75DD1195 pKa = 4.12TPTLSTVSPASLSNNATGFATGTSDD1220 pKa = 3.54PVIGADD1226 pKa = 3.78VPGTADD1232 pKa = 3.34LTGNIAGWNGTSVTYY1247 pKa = 10.24AASSLTSSGNTVYY1260 pKa = 10.98YY1261 pKa = 10.46AVSPVDD1267 pKa = 3.21TGVLYY1272 pKa = 10.4AYY1274 pKa = 9.19TSALPGTYY1282 pKa = 9.18TGGPGQSLIFTLTYY1296 pKa = 10.67DD1297 pKa = 3.15AAGNYY1302 pKa = 9.73AIDD1305 pKa = 3.86MNGKK1309 pKa = 9.64LDD1311 pKa = 4.21GPTQSFDD1318 pKa = 2.99ATFNQNIGGNQDD1330 pKa = 3.44YY1331 pKa = 11.25LLVTDD1336 pKa = 4.5TGALYY1341 pKa = 10.48KK1342 pKa = 10.26PGEE1345 pKa = 4.68SIPLGQTVIMTVDD1358 pKa = 3.18SSEE1361 pKa = 4.36GGVNSSMQGLAPDD1374 pKa = 4.02NQWVDD1379 pKa = 3.09GSEE1382 pKa = 4.31VIFFSYY1388 pKa = 10.85ASPIVSAQFSIDD1400 pKa = 3.21IQGGGSSNTVHH1411 pKa = 5.23WTVYY1415 pKa = 10.46GQDD1418 pKa = 3.41SLGNTTTQSGNTLFSDD1434 pKa = 3.97HH1435 pKa = 6.8VLTSIPTGLTAITRR1449 pKa = 11.84IDD1451 pKa = 3.68LSDD1454 pKa = 3.14TGGNGFRR1461 pKa = 11.84VNGSSIVEE1469 pKa = 4.42RR1470 pKa = 11.84IEE1472 pKa = 3.86EE1473 pKa = 4.25DD1474 pKa = 3.99PIATQFEE1481 pKa = 4.77VAVIDD1486 pKa = 4.23ADD1488 pKa = 3.75GDD1490 pKa = 3.62RR1491 pKa = 11.84AAAVLDD1497 pKa = 4.43VIFEE1501 pKa = 4.28PSLTGQLIVASNASDD1516 pKa = 3.62VAGSSAPYY1524 pKa = 8.22VTSGGAGVITGGAAADD1540 pKa = 3.49ILVGDD1545 pKa = 4.55EE1546 pKa = 4.11GRR1548 pKa = 11.84SNLVGKK1554 pKa = 9.56SVNLILILDD1563 pKa = 3.88SSGSMKK1569 pKa = 10.94ANIAFNDD1576 pKa = 3.73TNMSRR1581 pKa = 11.84MEE1583 pKa = 4.12ALKK1586 pKa = 10.79LAVNNLLVGLSLGAADD1602 pKa = 3.88HH1603 pKa = 6.53VRR1605 pKa = 11.84VQLVDD1610 pKa = 4.32FDD1612 pKa = 4.89SSASSLGIFDD1622 pKa = 5.3LKK1624 pKa = 11.08DD1625 pKa = 3.63GEE1627 pKa = 4.5LSAAQSAVNSLVAGGYY1643 pKa = 7.4TNYY1646 pKa = 10.41EE1647 pKa = 3.81AGLQVALNWVASGGVDD1663 pKa = 4.23APYY1666 pKa = 10.23TGANVINQVVFVSDD1680 pKa = 4.14GEE1682 pKa = 4.41PNSWLNGDD1690 pKa = 3.84STSLSNVTTDD1700 pKa = 3.33GSKK1703 pKa = 8.92STAVAQVLGSYY1714 pKa = 10.03NPSGHH1719 pKa = 6.75INDD1722 pKa = 4.28DD1723 pKa = 3.61RR1724 pKa = 11.84VSEE1727 pKa = 4.26VALLEE1732 pKa = 4.38STFGQIQAVGINVGGTALDD1751 pKa = 3.36ILNQVEE1757 pKa = 4.34GEE1759 pKa = 4.16LANVNPDD1766 pKa = 3.0VASNITTGEE1775 pKa = 3.81QLASVLADD1783 pKa = 4.0FNPSNQLHH1791 pKa = 6.9DD1792 pKa = 4.71AGSDD1796 pKa = 3.62HH1797 pKa = 6.84IVGGGGDD1804 pKa = 4.09DD1805 pKa = 5.64LIFGDD1810 pKa = 4.46VLYY1813 pKa = 10.47TDD1815 pKa = 4.42VLAATAGLTTNPGAGWQVFAALEE1838 pKa = 4.56GGDD1841 pKa = 4.19GPGSYY1846 pKa = 9.0ATWTRR1851 pKa = 11.84ADD1853 pKa = 3.49TLNYY1857 pKa = 9.51IRR1859 pKa = 11.84THH1861 pKa = 7.06AEE1863 pKa = 3.5EE1864 pKa = 4.2LAGEE1868 pKa = 4.24SGRR1871 pKa = 11.84ALGHH1875 pKa = 6.46DD1876 pKa = 4.61RR1877 pKa = 11.84IDD1879 pKa = 3.9AGDD1882 pKa = 3.97GNDD1885 pKa = 3.88LVFGQEE1891 pKa = 3.95GNDD1894 pKa = 3.63YY1895 pKa = 11.01ILGGKK1900 pKa = 9.59GNDD1903 pKa = 3.36VLAGGSGHH1911 pKa = 6.8DD1912 pKa = 3.61TLIGGEE1918 pKa = 4.23GDD1920 pKa = 3.64DD1921 pKa = 4.02VLIGGSGNDD1930 pKa = 4.03LLSGGLGKK1938 pKa = 10.32DD1939 pKa = 3.2SFVWNSGDD1947 pKa = 3.61SGIDD1951 pKa = 3.08HH1952 pKa = 6.01VTDD1955 pKa = 4.25FFIDD1959 pKa = 4.05GPNVSGGNSDD1969 pKa = 4.74VLDD1972 pKa = 4.41LSQLLVGEE1980 pKa = 4.44SASGNVLDD1988 pKa = 5.88DD1989 pKa = 3.73YY1990 pKa = 11.87LSFTFGASTAIDD2002 pKa = 3.51VRR2004 pKa = 11.84ATAGGAVVQQVVLDD2018 pKa = 4.22GVDD2021 pKa = 3.73LSSAAYY2027 pKa = 10.11YY2028 pKa = 10.89GSTDD2032 pKa = 3.26AATVIDD2038 pKa = 5.77GLLADD2043 pKa = 4.48NALKK2047 pKa = 10.72VDD2049 pKa = 3.98TVV2051 pKa = 3.3
MM1 pKa = 7.46LLDD4 pKa = 3.74EE5 pKa = 4.69TAGALDD11 pKa = 3.83PTIGFPTAGLSFSPEE26 pKa = 3.84FPDD29 pKa = 5.17PEE31 pKa = 4.69LEE33 pKa = 4.26PDD35 pKa = 3.62VTPEE39 pKa = 3.43PTAAVVPEE47 pKa = 5.09GPDD50 pKa = 3.15STPEE54 pKa = 3.7VEE56 pKa = 5.96IIHH59 pKa = 6.19QDD61 pKa = 3.01AEE63 pKa = 4.56GQVLGAPGVVDD74 pKa = 4.43EE75 pKa = 5.18AALATGSNAASNAEE89 pKa = 3.98QAFGRR94 pKa = 11.84LVVNSPDD101 pKa = 3.17GVAALQVQDD110 pKa = 5.3KK111 pKa = 10.43DD112 pKa = 4.33GNWVDD117 pKa = 3.62VTAGGTVQGVYY128 pKa = 10.92GSLVVDD134 pKa = 4.1AAGRR138 pKa = 11.84WVYY141 pKa = 9.37TLNGSTQDD149 pKa = 3.49HH150 pKa = 6.84GNPSAIGAADD160 pKa = 3.41QVGEE164 pKa = 4.24SFAVRR169 pKa = 11.84VFDD172 pKa = 3.59TDD174 pKa = 3.79GDD176 pKa = 4.01VSPVSTLSININDD189 pKa = 4.78DD190 pKa = 3.39GPNAVDD196 pKa = 4.21DD197 pKa = 4.56SNSIAEE203 pKa = 4.36DD204 pKa = 3.45TLTPISGNVLSNDD217 pKa = 3.44LHH219 pKa = 7.1PNGQPGADD227 pKa = 3.43TPISFVSWTSTAATYY242 pKa = 7.93GTFTDD247 pKa = 4.4TGNGTYY253 pKa = 9.97SYY255 pKa = 11.06SLNNANPAVQGLDD268 pKa = 3.1AGEE271 pKa = 4.39SLSEE275 pKa = 4.17TFSYY279 pKa = 10.45TMQDD283 pKa = 2.85ADD285 pKa = 4.37GDD287 pKa = 4.28TATATLTITITGSNDD302 pKa = 3.14VPVLTVDD309 pKa = 4.82PGNQGANDD317 pKa = 3.64RR318 pKa = 11.84VFEE321 pKa = 4.73AGLAQGSDD329 pKa = 3.03AASNSEE335 pKa = 4.07FASGTFTLSDD345 pKa = 3.83ADD347 pKa = 4.62GLDD350 pKa = 3.99DD351 pKa = 4.69LQSVTINGVTVAIGSLAGSQFIGAHH376 pKa = 5.67GSLTISAYY384 pKa = 9.04NTATGVASYY393 pKa = 10.35SYY395 pKa = 10.54EE396 pKa = 4.05LTSPTTDD403 pKa = 3.14GPGIEE408 pKa = 4.45SDD410 pKa = 3.76SFTLSVSDD418 pKa = 3.92GTASSAPASIVIEE431 pKa = 4.05IVDD434 pKa = 4.32DD435 pKa = 4.02VPHH438 pKa = 7.02AVDD441 pKa = 4.76DD442 pKa = 4.54SNATFASEE450 pKa = 4.08NQLTLIGNVLDD461 pKa = 4.73NDD463 pKa = 3.86VQGADD468 pKa = 4.2RR469 pKa = 11.84LPGGPIQPAVMQGTYY484 pKa = 8.95GTLTLNADD492 pKa = 3.47GSYY495 pKa = 10.73SYY497 pKa = 11.45LLDD500 pKa = 4.4PSDD503 pKa = 4.96ADD505 pKa = 3.97FLALGGGGMATEE517 pKa = 4.72TFTYY521 pKa = 9.52TLNDD525 pKa = 3.66ADD527 pKa = 4.77GDD529 pKa = 4.2SDD531 pKa = 4.0TANLVLKK538 pKa = 10.34VRR540 pKa = 11.84NLDD543 pKa = 3.59DD544 pKa = 3.6KK545 pKa = 11.81VQIKK549 pKa = 10.69DD550 pKa = 3.19LDD552 pKa = 3.81VRR554 pKa = 11.84GGEE557 pKa = 4.09HH558 pKa = 7.05RR559 pKa = 11.84VDD561 pKa = 3.86EE562 pKa = 5.62DD563 pKa = 3.93DD564 pKa = 4.84LAMGSDD570 pKa = 3.81PSKK573 pKa = 10.99EE574 pKa = 4.06STTVSGTFKK583 pKa = 10.86VQAPDD588 pKa = 3.17GLLNLTVGGIQVVLNGVVSGFPQSVTTPLGNQLTITGFDD627 pKa = 3.67ADD629 pKa = 5.73DD630 pKa = 5.42GILDD634 pKa = 3.99EE635 pKa = 5.97GIVSYY640 pKa = 10.82SYY642 pKa = 10.23TLLGSAQHH650 pKa = 6.46APGGGEE656 pKa = 3.91NQLTEE661 pKa = 4.26HH662 pKa = 6.43FAVLAEE668 pKa = 4.71DD669 pKa = 3.94VDD671 pKa = 4.15HH672 pKa = 7.23DD673 pKa = 4.26TDD675 pKa = 5.38LASLDD680 pKa = 3.55INIIDD685 pKa = 4.48DD686 pKa = 3.74VPHH689 pKa = 6.91AVDD692 pKa = 4.43DD693 pKa = 4.54SNSIAEE699 pKa = 4.47DD700 pKa = 3.42SVTPITGNVLSNDD713 pKa = 3.59LHH715 pKa = 7.1PNGQPGADD723 pKa = 3.37TPTSFVGWTSTAATYY738 pKa = 7.93GTFTDD743 pKa = 4.4TGNGTYY749 pKa = 9.97SYY751 pKa = 11.06SLNNANPAVQGLNTGEE767 pKa = 4.3SLSEE771 pKa = 3.96TFTYY775 pKa = 9.93TMRR778 pKa = 11.84DD779 pKa = 3.15ADD781 pKa = 4.37GDD783 pKa = 3.96TATAKK788 pKa = 9.58LTITINGADD797 pKa = 3.59DD798 pKa = 3.67HH799 pKa = 7.44ASVVTAAAYY808 pKa = 10.16GPDD811 pKa = 3.07ATVYY815 pKa = 9.82EE816 pKa = 4.62HH817 pKa = 7.16GLTSLADD824 pKa = 3.75TSEE827 pKa = 4.3STSGSFTVSASDD839 pKa = 4.65GIASVTVGGATFSLAQLQALAATNQLVSTGEE870 pKa = 4.06GTLTLTGYY878 pKa = 10.07SGSATAGTLSYY889 pKa = 10.89SYY891 pKa = 9.71TLNATIDD898 pKa = 3.87NDD900 pKa = 3.68SHH902 pKa = 8.18AGATDD907 pKa = 3.07THH909 pKa = 7.4FDD911 pKa = 3.68DD912 pKa = 4.41SLSISVLGVGGSTASDD928 pKa = 3.4EE929 pKa = 4.27LVVRR933 pKa = 11.84IIDD936 pKa = 3.88DD937 pKa = 3.8TPTAMNDD944 pKa = 3.43GPVGVTEE951 pKa = 4.44DD952 pKa = 3.62AVSNSVGGNVLTNDD966 pKa = 3.35SAGADD971 pKa = 3.61TPKK974 pKa = 11.09AFTTWAATGHH984 pKa = 7.0DD985 pKa = 3.49NTAAINALNSFGTLTLAAGGVYY1007 pKa = 10.41SFVLDD1012 pKa = 3.54NSLASVQALNASSHH1026 pKa = 5.97LSYY1029 pKa = 11.04DD1030 pKa = 3.22VYY1032 pKa = 11.01YY1033 pKa = 10.86TMQDD1037 pKa = 2.88ADD1039 pKa = 4.36GDD1041 pKa = 4.2SSSAKK1046 pKa = 9.41LTITISGADD1055 pKa = 3.43DD1056 pKa = 3.56HH1057 pKa = 7.52ASVVTAAAYY1066 pKa = 10.16GPDD1069 pKa = 3.07ATVYY1073 pKa = 9.82EE1074 pKa = 4.62HH1075 pKa = 7.16GLTSLADD1082 pKa = 3.68TSEE1085 pKa = 4.31TTSGSFTVSASDD1097 pKa = 4.65GIASVTVGGATFSFAQLQALATTNQQVSTGEE1128 pKa = 4.14GTLTLSNYY1136 pKa = 10.1SGSATAGTLSYY1147 pKa = 10.89SYY1149 pKa = 9.71TLNATIDD1156 pKa = 3.72NDD1158 pKa = 3.9SHH1160 pKa = 8.04VGATGTYY1167 pKa = 9.76FDD1169 pKa = 5.49DD1170 pKa = 3.94SLSISVMGVGGSAASDD1186 pKa = 3.59DD1187 pKa = 4.01LVVRR1191 pKa = 11.84IVDD1194 pKa = 3.75DD1195 pKa = 4.12TPTLSTVSPASLSNNATGFATGTSDD1220 pKa = 3.54PVIGADD1226 pKa = 3.78VPGTADD1232 pKa = 3.34LTGNIAGWNGTSVTYY1247 pKa = 10.24AASSLTSSGNTVYY1260 pKa = 10.98YY1261 pKa = 10.46AVSPVDD1267 pKa = 3.21TGVLYY1272 pKa = 10.4AYY1274 pKa = 9.19TSALPGTYY1282 pKa = 9.18TGGPGQSLIFTLTYY1296 pKa = 10.67DD1297 pKa = 3.15AAGNYY1302 pKa = 9.73AIDD1305 pKa = 3.86MNGKK1309 pKa = 9.64LDD1311 pKa = 4.21GPTQSFDD1318 pKa = 2.99ATFNQNIGGNQDD1330 pKa = 3.44YY1331 pKa = 11.25LLVTDD1336 pKa = 4.5TGALYY1341 pKa = 10.48KK1342 pKa = 10.26PGEE1345 pKa = 4.68SIPLGQTVIMTVDD1358 pKa = 3.18SSEE1361 pKa = 4.36GGVNSSMQGLAPDD1374 pKa = 4.02NQWVDD1379 pKa = 3.09GSEE1382 pKa = 4.31VIFFSYY1388 pKa = 10.85ASPIVSAQFSIDD1400 pKa = 3.21IQGGGSSNTVHH1411 pKa = 5.23WTVYY1415 pKa = 10.46GQDD1418 pKa = 3.41SLGNTTTQSGNTLFSDD1434 pKa = 3.97HH1435 pKa = 6.8VLTSIPTGLTAITRR1449 pKa = 11.84IDD1451 pKa = 3.68LSDD1454 pKa = 3.14TGGNGFRR1461 pKa = 11.84VNGSSIVEE1469 pKa = 4.42RR1470 pKa = 11.84IEE1472 pKa = 3.86EE1473 pKa = 4.25DD1474 pKa = 3.99PIATQFEE1481 pKa = 4.77VAVIDD1486 pKa = 4.23ADD1488 pKa = 3.75GDD1490 pKa = 3.62RR1491 pKa = 11.84AAAVLDD1497 pKa = 4.43VIFEE1501 pKa = 4.28PSLTGQLIVASNASDD1516 pKa = 3.62VAGSSAPYY1524 pKa = 8.22VTSGGAGVITGGAAADD1540 pKa = 3.49ILVGDD1545 pKa = 4.55EE1546 pKa = 4.11GRR1548 pKa = 11.84SNLVGKK1554 pKa = 9.56SVNLILILDD1563 pKa = 3.88SSGSMKK1569 pKa = 10.94ANIAFNDD1576 pKa = 3.73TNMSRR1581 pKa = 11.84MEE1583 pKa = 4.12ALKK1586 pKa = 10.79LAVNNLLVGLSLGAADD1602 pKa = 3.88HH1603 pKa = 6.53VRR1605 pKa = 11.84VQLVDD1610 pKa = 4.32FDD1612 pKa = 4.89SSASSLGIFDD1622 pKa = 5.3LKK1624 pKa = 11.08DD1625 pKa = 3.63GEE1627 pKa = 4.5LSAAQSAVNSLVAGGYY1643 pKa = 7.4TNYY1646 pKa = 10.41EE1647 pKa = 3.81AGLQVALNWVASGGVDD1663 pKa = 4.23APYY1666 pKa = 10.23TGANVINQVVFVSDD1680 pKa = 4.14GEE1682 pKa = 4.41PNSWLNGDD1690 pKa = 3.84STSLSNVTTDD1700 pKa = 3.33GSKK1703 pKa = 8.92STAVAQVLGSYY1714 pKa = 10.03NPSGHH1719 pKa = 6.75INDD1722 pKa = 4.28DD1723 pKa = 3.61RR1724 pKa = 11.84VSEE1727 pKa = 4.26VALLEE1732 pKa = 4.38STFGQIQAVGINVGGTALDD1751 pKa = 3.36ILNQVEE1757 pKa = 4.34GEE1759 pKa = 4.16LANVNPDD1766 pKa = 3.0VASNITTGEE1775 pKa = 3.81QLASVLADD1783 pKa = 4.0FNPSNQLHH1791 pKa = 6.9DD1792 pKa = 4.71AGSDD1796 pKa = 3.62HH1797 pKa = 6.84IVGGGGDD1804 pKa = 4.09DD1805 pKa = 5.64LIFGDD1810 pKa = 4.46VLYY1813 pKa = 10.47TDD1815 pKa = 4.42VLAATAGLTTNPGAGWQVFAALEE1838 pKa = 4.56GGDD1841 pKa = 4.19GPGSYY1846 pKa = 9.0ATWTRR1851 pKa = 11.84ADD1853 pKa = 3.49TLNYY1857 pKa = 9.51IRR1859 pKa = 11.84THH1861 pKa = 7.06AEE1863 pKa = 3.5EE1864 pKa = 4.2LAGEE1868 pKa = 4.24SGRR1871 pKa = 11.84ALGHH1875 pKa = 6.46DD1876 pKa = 4.61RR1877 pKa = 11.84IDD1879 pKa = 3.9AGDD1882 pKa = 3.97GNDD1885 pKa = 3.88LVFGQEE1891 pKa = 3.95GNDD1894 pKa = 3.63YY1895 pKa = 11.01ILGGKK1900 pKa = 9.59GNDD1903 pKa = 3.36VLAGGSGHH1911 pKa = 6.8DD1912 pKa = 3.61TLIGGEE1918 pKa = 4.23GDD1920 pKa = 3.64DD1921 pKa = 4.02VLIGGSGNDD1930 pKa = 4.03LLSGGLGKK1938 pKa = 10.32DD1939 pKa = 3.2SFVWNSGDD1947 pKa = 3.61SGIDD1951 pKa = 3.08HH1952 pKa = 6.01VTDD1955 pKa = 4.25FFIDD1959 pKa = 4.05GPNVSGGNSDD1969 pKa = 4.74VLDD1972 pKa = 4.41LSQLLVGEE1980 pKa = 4.44SASGNVLDD1988 pKa = 5.88DD1989 pKa = 3.73YY1990 pKa = 11.87LSFTFGASTAIDD2002 pKa = 3.51VRR2004 pKa = 11.84ATAGGAVVQQVVLDD2018 pKa = 4.22GVDD2021 pKa = 3.73LSSAAYY2027 pKa = 10.11YY2028 pKa = 10.89GSTDD2032 pKa = 3.26AATVIDD2038 pKa = 5.77GLLADD2043 pKa = 4.48NALKK2047 pKa = 10.72VDD2049 pKa = 3.98TVV2051 pKa = 3.3
Molecular weight: 208.09 kDa
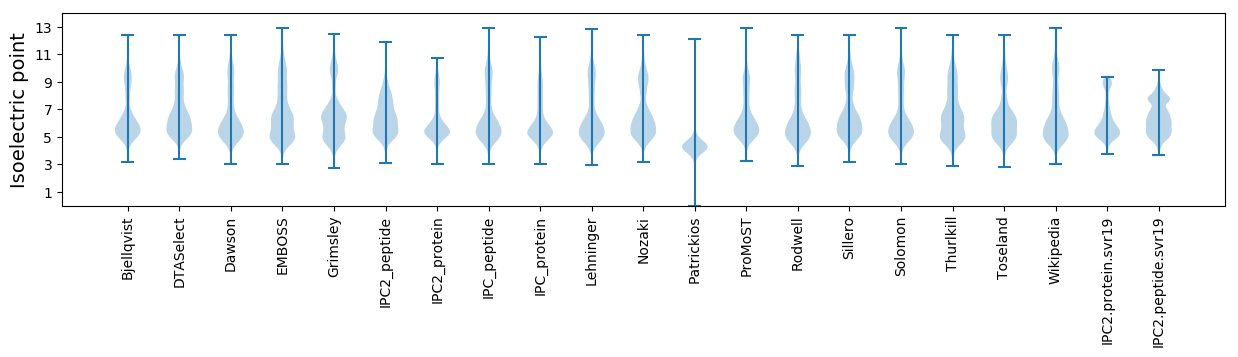
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1YGA6|A0A0A1YGA6_9PSED Uncharacterized protein OS=Pseudomonas taeanensis MS-3 OX=1395571 GN=TMS3_0120440 PE=4 SV=1
MM1 pKa = 7.78PKK3 pKa = 10.48LPTALQHH10 pKa = 5.63WRR12 pKa = 11.84SWWLTPLIGLVGGYY26 pKa = 8.99LASLIGWPLPWIIGSLLAVIAARR49 pKa = 11.84CSGWRR54 pKa = 11.84VTEE57 pKa = 4.12LPGGRR62 pKa = 11.84QTGQWLVASGIGLHH76 pKa = 5.39FTGAVMAQVLSHH88 pKa = 6.83FPLILAGALGTLLLSLIGIALLRR111 pKa = 11.84RR112 pKa = 11.84AGVDD116 pKa = 2.61RR117 pKa = 11.84ATAFFASMPGGASEE131 pKa = 4.42MVNLAQRR138 pKa = 11.84HH139 pKa = 5.45DD140 pKa = 3.65AQIARR145 pKa = 11.84VAAAHH150 pKa = 5.84SLRR153 pKa = 11.84LLLVVLLIPALFTWSLAPIQAPAPAPVDD181 pKa = 3.92WYY183 pKa = 9.94WLALLLPGGALLALLWKK200 pKa = 10.33KK201 pKa = 10.12LRR203 pKa = 11.84QPNPWMLGPLTVCAAASVTFDD224 pKa = 4.07LQLGLPPGLGQVGQWLIGCALGCHH248 pKa = 6.53FDD250 pKa = 3.34RR251 pKa = 11.84AFFRR255 pKa = 11.84SAPGFLARR263 pKa = 11.84ILLFSLLAMLGAALLAQGLSWLASQDD289 pKa = 3.39GTSLMLGMMPGGITEE304 pKa = 4.67LCLTAEE310 pKa = 4.39ALQLSVALVTALQVLRR326 pKa = 11.84LFLVMFLAEE335 pKa = 4.03PLFRR339 pKa = 11.84LWQRR343 pKa = 11.84SS344 pKa = 3.29
MM1 pKa = 7.78PKK3 pKa = 10.48LPTALQHH10 pKa = 5.63WRR12 pKa = 11.84SWWLTPLIGLVGGYY26 pKa = 8.99LASLIGWPLPWIIGSLLAVIAARR49 pKa = 11.84CSGWRR54 pKa = 11.84VTEE57 pKa = 4.12LPGGRR62 pKa = 11.84QTGQWLVASGIGLHH76 pKa = 5.39FTGAVMAQVLSHH88 pKa = 6.83FPLILAGALGTLLLSLIGIALLRR111 pKa = 11.84RR112 pKa = 11.84AGVDD116 pKa = 2.61RR117 pKa = 11.84ATAFFASMPGGASEE131 pKa = 4.42MVNLAQRR138 pKa = 11.84HH139 pKa = 5.45DD140 pKa = 3.65AQIARR145 pKa = 11.84VAAAHH150 pKa = 5.84SLRR153 pKa = 11.84LLLVVLLIPALFTWSLAPIQAPAPAPVDD181 pKa = 3.92WYY183 pKa = 9.94WLALLLPGGALLALLWKK200 pKa = 10.33KK201 pKa = 10.12LRR203 pKa = 11.84QPNPWMLGPLTVCAAASVTFDD224 pKa = 4.07LQLGLPPGLGQVGQWLIGCALGCHH248 pKa = 6.53FDD250 pKa = 3.34RR251 pKa = 11.84AFFRR255 pKa = 11.84SAPGFLARR263 pKa = 11.84ILLFSLLAMLGAALLAQGLSWLASQDD289 pKa = 3.39GTSLMLGMMPGGITEE304 pKa = 4.67LCLTAEE310 pKa = 4.39ALQLSVALVTALQVLRR326 pKa = 11.84LFLVMFLAEE335 pKa = 4.03PLFRR339 pKa = 11.84LWQRR343 pKa = 11.84SS344 pKa = 3.29
Molecular weight: 36.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1480677 |
35 |
2430 |
323.8 |
35.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.192 ± 0.046 | 1.062 ± 0.014 |
5.182 ± 0.027 | 5.94 ± 0.032 |
3.635 ± 0.022 | 7.882 ± 0.034 |
2.307 ± 0.019 | 4.741 ± 0.027 |
3.292 ± 0.027 | 12.282 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.284 ± 0.02 | 2.876 ± 0.021 |
4.764 ± 0.025 | 4.875 ± 0.036 |
6.466 ± 0.038 | 5.867 ± 0.025 |
4.507 ± 0.025 | 6.914 ± 0.032 |
1.42 ± 0.015 | 2.513 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
