
Parasedimentitalea marina
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Parasedimentitalea
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
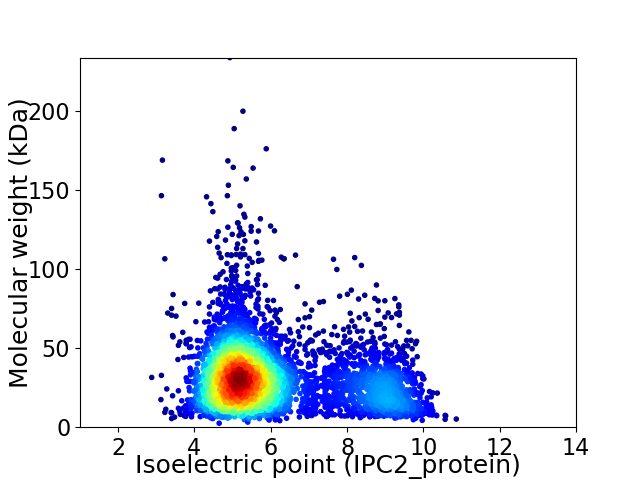
Virtual 2D-PAGE plot for 4642 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
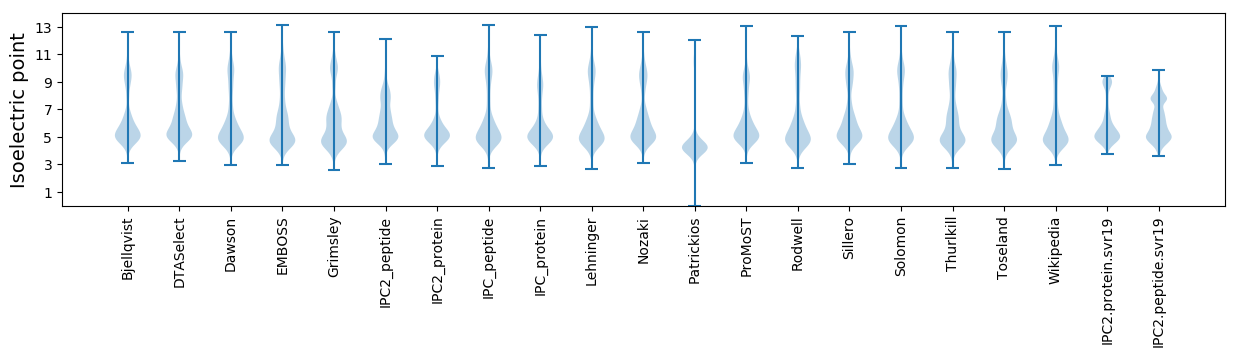
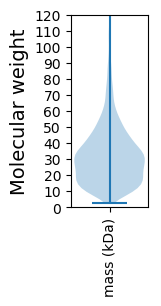
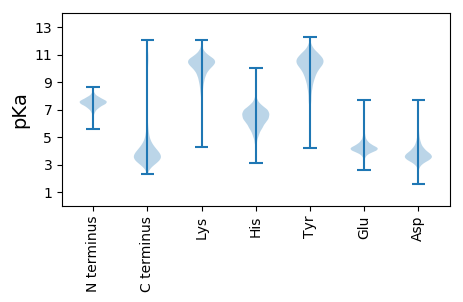
Protein with the lowest isoelectric point:
>tr|A0A3T0N3V4|A0A3T0N3V4_9RHOB MerR family transcriptional regulator OS=Parasedimentitalea marina OX=2483033 GN=EBB79_12775 PE=4 SV=1
MM1 pKa = 7.07ATYY4 pKa = 10.28VPHH7 pKa = 6.96ASYY10 pKa = 11.07RR11 pKa = 11.84LYY13 pKa = 11.18DD14 pKa = 3.36FAGRR18 pKa = 11.84LFAAEE23 pKa = 4.25EE24 pKa = 4.28VEE26 pKa = 4.41ITQTSNGVDD35 pKa = 3.12IQFLGFDD42 pKa = 3.32RR43 pKa = 11.84RR44 pKa = 11.84TGEE47 pKa = 4.17VKK49 pKa = 10.73VKK51 pKa = 10.8GGMRR55 pKa = 11.84LSGDD59 pKa = 3.38NLVVDD64 pKa = 3.81NGEE67 pKa = 4.16IVSGSIGTVQFFAFEE82 pKa = 3.89FEE84 pKa = 4.72YY85 pKa = 11.0YY86 pKa = 10.46AGRR89 pKa = 11.84LEE91 pKa = 4.01SLNWDD96 pKa = 3.33GTYY99 pKa = 10.55LQNFLSEE106 pKa = 4.34TTDD109 pKa = 3.45GNFSPFYY116 pKa = 10.38DD117 pKa = 3.86ALTGAVDD124 pKa = 3.99LVRR127 pKa = 11.84GISSDD132 pKa = 3.71DD133 pKa = 3.95NYY135 pKa = 11.75DD136 pKa = 3.1HH137 pKa = 7.45SEE139 pKa = 4.41YY140 pKa = 11.32YY141 pKa = 10.54DD142 pKa = 3.3ATDD145 pKa = 2.83IHH147 pKa = 7.79LGAGNDD153 pKa = 3.96SYY155 pKa = 10.72THH157 pKa = 6.45PRR159 pKa = 11.84LTEE162 pKa = 3.84MSPLIIDD169 pKa = 3.83GGEE172 pKa = 3.93GHH174 pKa = 7.62DD175 pKa = 4.09NFQAGIFGSYY185 pKa = 10.11EE186 pKa = 3.67PLLIDD191 pKa = 3.26VAKK194 pKa = 10.8NLFVDD199 pKa = 4.73EE200 pKa = 4.72YY201 pKa = 10.83GVEE204 pKa = 3.9HH205 pKa = 6.11VVRR208 pKa = 11.84NFEE211 pKa = 4.15FFQGGRR217 pKa = 11.84GNDD220 pKa = 3.7TLIGAMDD227 pKa = 4.88RR228 pKa = 11.84PNQFYY233 pKa = 10.32GYY235 pKa = 9.5QGSDD239 pKa = 3.36HH240 pKa = 6.97ISGGNHH246 pKa = 7.23DD247 pKa = 5.54DD248 pKa = 3.8YY249 pKa = 11.72LAGGSGLDD257 pKa = 3.48TILAYY262 pKa = 10.42GGDD265 pKa = 4.27DD266 pKa = 4.0YY267 pKa = 11.7ILGSSHH273 pKa = 7.67DD274 pKa = 4.18YY275 pKa = 11.34VDD277 pKa = 4.16GGDD280 pKa = 4.27GIDD283 pKa = 3.28TFGYY287 pKa = 10.24RR288 pKa = 11.84LIAGYY293 pKa = 10.51VYY295 pKa = 10.36IDD297 pKa = 5.36LEE299 pKa = 4.18NQEE302 pKa = 4.92DD303 pKa = 3.84NRR305 pKa = 11.84GDD307 pKa = 3.79AIYY310 pKa = 9.49TVLVNIEE317 pKa = 4.17NLNGSNYY324 pKa = 9.03YY325 pKa = 10.98DD326 pKa = 4.36VMLGDD331 pKa = 4.59ANDD334 pKa = 3.71NVIEE338 pKa = 4.29TFGYY342 pKa = 10.41RR343 pKa = 11.84DD344 pKa = 3.44ILHH347 pKa = 6.67GRR349 pKa = 11.84DD350 pKa = 3.83GNDD353 pKa = 3.13TLDD356 pKa = 3.76AGEE359 pKa = 4.96GNDD362 pKa = 4.49DD363 pKa = 3.83LSGGTGANLLIGGAGADD380 pKa = 3.24NFIFEE385 pKa = 4.64TQNATDD391 pKa = 4.44TIADD395 pKa = 3.86FEE397 pKa = 4.69VGIDD401 pKa = 3.66SLDD404 pKa = 3.74FSALTSQADD413 pKa = 4.31FINFEE418 pKa = 4.81LNGMSYY424 pKa = 11.01SRR426 pKa = 11.84ISCFGDD432 pKa = 3.78FKK434 pKa = 11.09LGNVSLRR441 pKa = 11.84FVQSQEE447 pKa = 3.85DD448 pKa = 3.84VEE450 pKa = 5.07IYY452 pKa = 10.7LDD454 pKa = 3.78PSLGGEE460 pKa = 4.27SASPLITLIDD470 pKa = 3.6IGIHH474 pKa = 6.19DD475 pKa = 6.15LIVDD479 pKa = 3.84DD480 pKa = 6.89FIFF483 pKa = 4.49
MM1 pKa = 7.07ATYY4 pKa = 10.28VPHH7 pKa = 6.96ASYY10 pKa = 11.07RR11 pKa = 11.84LYY13 pKa = 11.18DD14 pKa = 3.36FAGRR18 pKa = 11.84LFAAEE23 pKa = 4.25EE24 pKa = 4.28VEE26 pKa = 4.41ITQTSNGVDD35 pKa = 3.12IQFLGFDD42 pKa = 3.32RR43 pKa = 11.84RR44 pKa = 11.84TGEE47 pKa = 4.17VKK49 pKa = 10.73VKK51 pKa = 10.8GGMRR55 pKa = 11.84LSGDD59 pKa = 3.38NLVVDD64 pKa = 3.81NGEE67 pKa = 4.16IVSGSIGTVQFFAFEE82 pKa = 3.89FEE84 pKa = 4.72YY85 pKa = 11.0YY86 pKa = 10.46AGRR89 pKa = 11.84LEE91 pKa = 4.01SLNWDD96 pKa = 3.33GTYY99 pKa = 10.55LQNFLSEE106 pKa = 4.34TTDD109 pKa = 3.45GNFSPFYY116 pKa = 10.38DD117 pKa = 3.86ALTGAVDD124 pKa = 3.99LVRR127 pKa = 11.84GISSDD132 pKa = 3.71DD133 pKa = 3.95NYY135 pKa = 11.75DD136 pKa = 3.1HH137 pKa = 7.45SEE139 pKa = 4.41YY140 pKa = 11.32YY141 pKa = 10.54DD142 pKa = 3.3ATDD145 pKa = 2.83IHH147 pKa = 7.79LGAGNDD153 pKa = 3.96SYY155 pKa = 10.72THH157 pKa = 6.45PRR159 pKa = 11.84LTEE162 pKa = 3.84MSPLIIDD169 pKa = 3.83GGEE172 pKa = 3.93GHH174 pKa = 7.62DD175 pKa = 4.09NFQAGIFGSYY185 pKa = 10.11EE186 pKa = 3.67PLLIDD191 pKa = 3.26VAKK194 pKa = 10.8NLFVDD199 pKa = 4.73EE200 pKa = 4.72YY201 pKa = 10.83GVEE204 pKa = 3.9HH205 pKa = 6.11VVRR208 pKa = 11.84NFEE211 pKa = 4.15FFQGGRR217 pKa = 11.84GNDD220 pKa = 3.7TLIGAMDD227 pKa = 4.88RR228 pKa = 11.84PNQFYY233 pKa = 10.32GYY235 pKa = 9.5QGSDD239 pKa = 3.36HH240 pKa = 6.97ISGGNHH246 pKa = 7.23DD247 pKa = 5.54DD248 pKa = 3.8YY249 pKa = 11.72LAGGSGLDD257 pKa = 3.48TILAYY262 pKa = 10.42GGDD265 pKa = 4.27DD266 pKa = 4.0YY267 pKa = 11.7ILGSSHH273 pKa = 7.67DD274 pKa = 4.18YY275 pKa = 11.34VDD277 pKa = 4.16GGDD280 pKa = 4.27GIDD283 pKa = 3.28TFGYY287 pKa = 10.24RR288 pKa = 11.84LIAGYY293 pKa = 10.51VYY295 pKa = 10.36IDD297 pKa = 5.36LEE299 pKa = 4.18NQEE302 pKa = 4.92DD303 pKa = 3.84NRR305 pKa = 11.84GDD307 pKa = 3.79AIYY310 pKa = 9.49TVLVNIEE317 pKa = 4.17NLNGSNYY324 pKa = 9.03YY325 pKa = 10.98DD326 pKa = 4.36VMLGDD331 pKa = 4.59ANDD334 pKa = 3.71NVIEE338 pKa = 4.29TFGYY342 pKa = 10.41RR343 pKa = 11.84DD344 pKa = 3.44ILHH347 pKa = 6.67GRR349 pKa = 11.84DD350 pKa = 3.83GNDD353 pKa = 3.13TLDD356 pKa = 3.76AGEE359 pKa = 4.96GNDD362 pKa = 4.49DD363 pKa = 3.83LSGGTGANLLIGGAGADD380 pKa = 3.24NFIFEE385 pKa = 4.64TQNATDD391 pKa = 4.44TIADD395 pKa = 3.86FEE397 pKa = 4.69VGIDD401 pKa = 3.66SLDD404 pKa = 3.74FSALTSQADD413 pKa = 4.31FINFEE418 pKa = 4.81LNGMSYY424 pKa = 11.01SRR426 pKa = 11.84ISCFGDD432 pKa = 3.78FKK434 pKa = 11.09LGNVSLRR441 pKa = 11.84FVQSQEE447 pKa = 3.85DD448 pKa = 3.84VEE450 pKa = 5.07IYY452 pKa = 10.7LDD454 pKa = 3.78PSLGGEE460 pKa = 4.27SASPLITLIDD470 pKa = 3.6IGIHH474 pKa = 6.19DD475 pKa = 6.15LIVDD479 pKa = 3.84DD480 pKa = 6.89FIFF483 pKa = 4.49
Molecular weight: 52.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3T0MYF2|A0A3T0MYF2_9RHOB Uncharacterized protein OS=Parasedimentitalea marina OX=2483033 GN=EBB79_02025 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 5.39GRR39 pKa = 11.84KK40 pKa = 8.9EE41 pKa = 3.73LSAA44 pKa = 4.94
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 5.39GRR39 pKa = 11.84KK40 pKa = 8.9EE41 pKa = 3.73LSAA44 pKa = 4.94
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1420733 |
26 |
2158 |
306.1 |
33.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.253 ± 0.045 | 0.97 ± 0.012 |
6.054 ± 0.029 | 5.55 ± 0.037 |
3.842 ± 0.024 | 8.375 ± 0.035 |
2.115 ± 0.017 | 5.43 ± 0.023 |
3.504 ± 0.032 | 10.14 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.854 ± 0.016 | 3.007 ± 0.019 |
4.822 ± 0.028 | 3.719 ± 0.021 |
5.949 ± 0.032 | 5.893 ± 0.032 |
5.631 ± 0.025 | 7.182 ± 0.027 |
1.389 ± 0.015 | 2.321 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
