
Perkinsus sp. BL_2016
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Perkinsozoa; Perkinsea; Perkinsida; Perkinsidae; Perkinsus; unclassified Perkinsus
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
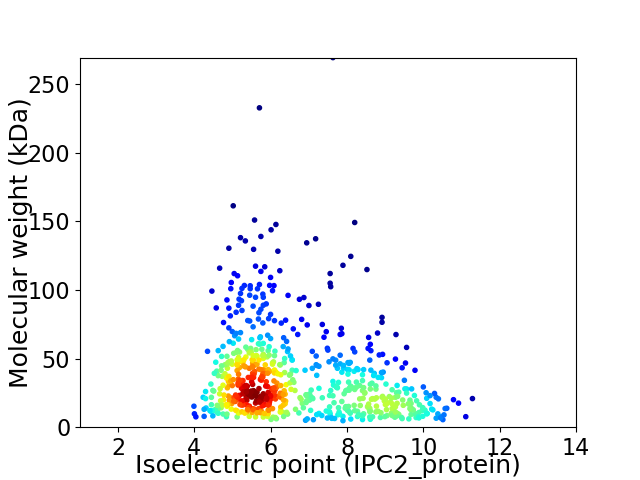
Virtual 2D-PAGE plot for 691 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
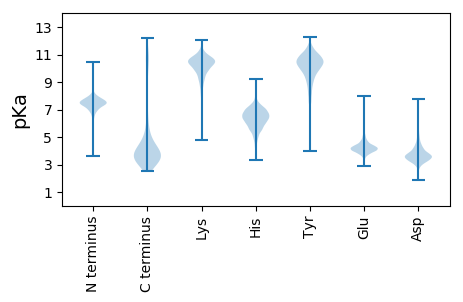
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D8XLH6|A0A4D8XLH6_9ALVE Uncharacterized protein OS=Perkinsus sp. BL_2016 OX=2494336 GN=C9890_0299 PE=4 SV=1
MM1 pKa = 6.4TTCNSYY7 pKa = 11.11CSDD10 pKa = 3.5FVSGSNCQLWLPIPLCQGTDD30 pKa = 3.3VRR32 pKa = 11.84CGYY35 pKa = 10.33DD36 pKa = 2.93SCRR39 pKa = 11.84VNSRR43 pKa = 11.84PSRR46 pKa = 11.84QIVAKK51 pKa = 10.05SATSSHH57 pKa = 6.11SVTLTRR63 pKa = 11.84IIVSGSYY70 pKa = 7.89PTSTPTSPTMMTTIIPEE87 pKa = 4.5GSATNSHH94 pKa = 6.28SADD97 pKa = 3.16LTQIVVDD104 pKa = 5.13GPHH107 pKa = 7.04PGSTTTTTASSEE119 pKa = 4.41TTPEE123 pKa = 3.71APMSVSVVPITSQDD137 pKa = 3.33PTATASTTEE146 pKa = 4.13SEE148 pKa = 4.54APMSVSAVPISSSDD162 pKa = 3.06PSMAYY167 pKa = 10.34VSVAEE172 pKa = 4.21STEE175 pKa = 4.61DD176 pKa = 3.8IADD179 pKa = 3.76SDD181 pKa = 4.1PLEE184 pKa = 4.39NCISRR189 pKa = 11.84AEE191 pKa = 3.94SDD193 pKa = 3.45EE194 pKa = 5.54NIFLWAEE201 pKa = 3.96WPSIPSEE208 pKa = 3.95ANSWINYY215 pKa = 6.72YY216 pKa = 11.08SSMASFIDD224 pKa = 4.32GNCARR229 pKa = 11.84LKK231 pKa = 8.44VTRR234 pKa = 11.84LILRR238 pKa = 11.84VTDD241 pKa = 4.08PLKK244 pKa = 10.64GAWWPPLDD252 pKa = 3.73SPMYY256 pKa = 10.01TSLLRR261 pKa = 11.84NIPEE265 pKa = 3.86EE266 pKa = 4.36TEE268 pKa = 4.36VYY270 pKa = 9.97VYY272 pKa = 10.07PYY274 pKa = 11.67VMDD277 pKa = 3.95QWNQDD282 pKa = 2.57QWTCFSPSNTSVLEE296 pKa = 4.08GAFEE300 pKa = 4.4FAKK303 pKa = 10.22RR304 pKa = 11.84WNDD307 pKa = 3.13FLTQQTSSLRR317 pKa = 11.84FVGIVLDD324 pKa = 3.83AEE326 pKa = 4.43EE327 pKa = 4.56FSGGRR332 pKa = 11.84NPEE335 pKa = 3.69FRR337 pKa = 11.84AQLLNSTQLKK347 pKa = 9.49QKK349 pKa = 10.7YY350 pKa = 7.71SLKK353 pKa = 10.69LGLSLGYY360 pKa = 10.37QSGPQMAAWDD370 pKa = 4.04SVVDD374 pKa = 3.73AFYY377 pKa = 10.98LQFYY381 pKa = 9.85DD382 pKa = 4.15YY383 pKa = 10.85YY384 pKa = 8.26YY385 pKa = 10.46TPYY388 pKa = 10.8VDD390 pKa = 4.26ATADD394 pKa = 3.91SPFLLYY400 pKa = 11.14LNDD403 pKa = 3.99PASLVGFTLDD413 pKa = 3.23TVLEE417 pKa = 4.71GKK419 pKa = 9.97SEE421 pKa = 4.12DD422 pKa = 3.46QTKK425 pKa = 10.39YY426 pKa = 9.65GPKK429 pKa = 9.97VSVMWSMQNLGTGCIFPLNDD449 pKa = 3.7GSCGVNFEE457 pKa = 5.28FGAGWSAQAFNEE469 pKa = 3.94YY470 pKa = 10.42LKK472 pKa = 9.9EE473 pKa = 3.87FRR475 pKa = 11.84MRR477 pKa = 11.84SPNLGYY483 pKa = 10.24KK484 pKa = 9.07PQGIFQYY491 pKa = 11.16SFLPQSWFLTT501 pKa = 4.06
MM1 pKa = 6.4TTCNSYY7 pKa = 11.11CSDD10 pKa = 3.5FVSGSNCQLWLPIPLCQGTDD30 pKa = 3.3VRR32 pKa = 11.84CGYY35 pKa = 10.33DD36 pKa = 2.93SCRR39 pKa = 11.84VNSRR43 pKa = 11.84PSRR46 pKa = 11.84QIVAKK51 pKa = 10.05SATSSHH57 pKa = 6.11SVTLTRR63 pKa = 11.84IIVSGSYY70 pKa = 7.89PTSTPTSPTMMTTIIPEE87 pKa = 4.5GSATNSHH94 pKa = 6.28SADD97 pKa = 3.16LTQIVVDD104 pKa = 5.13GPHH107 pKa = 7.04PGSTTTTTASSEE119 pKa = 4.41TTPEE123 pKa = 3.71APMSVSVVPITSQDD137 pKa = 3.33PTATASTTEE146 pKa = 4.13SEE148 pKa = 4.54APMSVSAVPISSSDD162 pKa = 3.06PSMAYY167 pKa = 10.34VSVAEE172 pKa = 4.21STEE175 pKa = 4.61DD176 pKa = 3.8IADD179 pKa = 3.76SDD181 pKa = 4.1PLEE184 pKa = 4.39NCISRR189 pKa = 11.84AEE191 pKa = 3.94SDD193 pKa = 3.45EE194 pKa = 5.54NIFLWAEE201 pKa = 3.96WPSIPSEE208 pKa = 3.95ANSWINYY215 pKa = 6.72YY216 pKa = 11.08SSMASFIDD224 pKa = 4.32GNCARR229 pKa = 11.84LKK231 pKa = 8.44VTRR234 pKa = 11.84LILRR238 pKa = 11.84VTDD241 pKa = 4.08PLKK244 pKa = 10.64GAWWPPLDD252 pKa = 3.73SPMYY256 pKa = 10.01TSLLRR261 pKa = 11.84NIPEE265 pKa = 3.86EE266 pKa = 4.36TEE268 pKa = 4.36VYY270 pKa = 9.97VYY272 pKa = 10.07PYY274 pKa = 11.67VMDD277 pKa = 3.95QWNQDD282 pKa = 2.57QWTCFSPSNTSVLEE296 pKa = 4.08GAFEE300 pKa = 4.4FAKK303 pKa = 10.22RR304 pKa = 11.84WNDD307 pKa = 3.13FLTQQTSSLRR317 pKa = 11.84FVGIVLDD324 pKa = 3.83AEE326 pKa = 4.43EE327 pKa = 4.56FSGGRR332 pKa = 11.84NPEE335 pKa = 3.69FRR337 pKa = 11.84AQLLNSTQLKK347 pKa = 9.49QKK349 pKa = 10.7YY350 pKa = 7.71SLKK353 pKa = 10.69LGLSLGYY360 pKa = 10.37QSGPQMAAWDD370 pKa = 4.04SVVDD374 pKa = 3.73AFYY377 pKa = 10.98LQFYY381 pKa = 9.85DD382 pKa = 4.15YY383 pKa = 10.85YY384 pKa = 8.26YY385 pKa = 10.46TPYY388 pKa = 10.8VDD390 pKa = 4.26ATADD394 pKa = 3.91SPFLLYY400 pKa = 11.14LNDD403 pKa = 3.99PASLVGFTLDD413 pKa = 3.23TVLEE417 pKa = 4.71GKK419 pKa = 9.97SEE421 pKa = 4.12DD422 pKa = 3.46QTKK425 pKa = 10.39YY426 pKa = 9.65GPKK429 pKa = 9.97VSVMWSMQNLGTGCIFPLNDD449 pKa = 3.7GSCGVNFEE457 pKa = 5.28FGAGWSAQAFNEE469 pKa = 3.94YY470 pKa = 10.42LKK472 pKa = 9.9EE473 pKa = 3.87FRR475 pKa = 11.84MRR477 pKa = 11.84SPNLGYY483 pKa = 10.24KK484 pKa = 9.07PQGIFQYY491 pKa = 11.16SFLPQSWFLTT501 pKa = 4.06
Molecular weight: 55.31 kDa
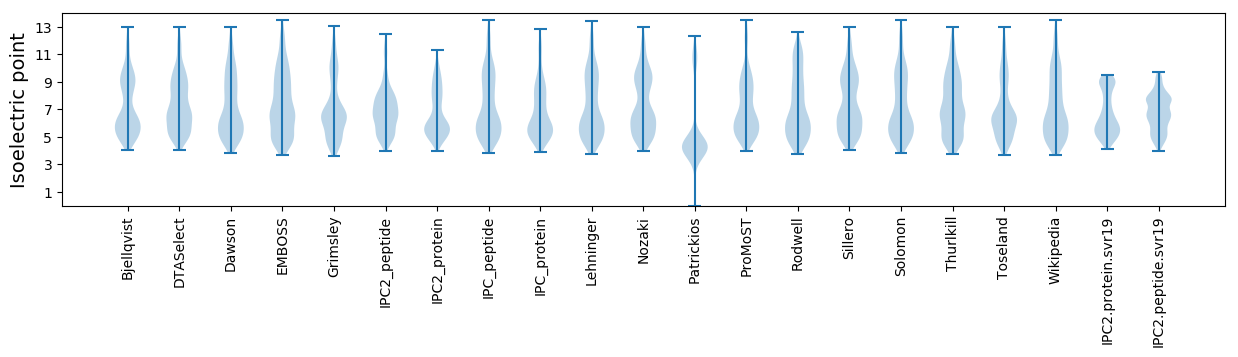
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D8XPI3|A0A4D8XPI3_9ALVE Uncharacterized protein OS=Perkinsus sp. BL_2016 OX=2494336 GN=C9890_0706 PE=4 SV=1
SS1 pKa = 6.82SGRR4 pKa = 11.84SLRR7 pKa = 11.84SRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ACRR16 pKa = 11.84RR17 pKa = 11.84ARR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 9.83PDD23 pKa = 2.82VHH25 pKa = 7.53LPLGRR30 pKa = 11.84TGMARR35 pKa = 11.84RR36 pKa = 11.84PPAARR41 pKa = 11.84PRR43 pKa = 11.84WHH45 pKa = 7.15RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84SLQRR52 pKa = 11.84TPLRR56 pKa = 11.84GHH58 pKa = 6.59RR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84SPPRR65 pKa = 11.84HH66 pKa = 5.49LAAYY70 pKa = 7.2DD71 pKa = 3.76HH72 pKa = 7.08PPAPVFALHH81 pKa = 5.32SHH83 pKa = 5.34RR84 pKa = 11.84HH85 pKa = 4.73RR86 pKa = 11.84LRR88 pKa = 11.84RR89 pKa = 11.84LASGARR95 pKa = 11.84RR96 pKa = 11.84TRR98 pKa = 11.84PHH100 pKa = 6.79RR101 pKa = 11.84RR102 pKa = 11.84PNNLPGFRR110 pKa = 11.84PLRR113 pKa = 11.84PGPRR117 pKa = 11.84TGNVPRR123 pKa = 11.84MRR125 pKa = 11.84RR126 pKa = 11.84RR127 pKa = 11.84HH128 pKa = 5.89APPSQPSRR136 pKa = 11.84PRR138 pKa = 11.84RR139 pKa = 11.84IAPRR143 pKa = 11.84TTQRR147 pKa = 11.84RR148 pKa = 11.84QSPRR152 pKa = 11.84SRR154 pKa = 11.84GPPRR158 pKa = 11.84SRR160 pKa = 11.84PTPRR164 pKa = 11.84HH165 pKa = 5.64PPRR168 pKa = 11.84SRR170 pKa = 11.84SPRR173 pKa = 11.84PLPTT177 pKa = 3.53
SS1 pKa = 6.82SGRR4 pKa = 11.84SLRR7 pKa = 11.84SRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ACRR16 pKa = 11.84RR17 pKa = 11.84ARR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 9.83PDD23 pKa = 2.82VHH25 pKa = 7.53LPLGRR30 pKa = 11.84TGMARR35 pKa = 11.84RR36 pKa = 11.84PPAARR41 pKa = 11.84PRR43 pKa = 11.84WHH45 pKa = 7.15RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84SLQRR52 pKa = 11.84TPLRR56 pKa = 11.84GHH58 pKa = 6.59RR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84SPPRR65 pKa = 11.84HH66 pKa = 5.49LAAYY70 pKa = 7.2DD71 pKa = 3.76HH72 pKa = 7.08PPAPVFALHH81 pKa = 5.32SHH83 pKa = 5.34RR84 pKa = 11.84HH85 pKa = 4.73RR86 pKa = 11.84LRR88 pKa = 11.84RR89 pKa = 11.84LASGARR95 pKa = 11.84RR96 pKa = 11.84TRR98 pKa = 11.84PHH100 pKa = 6.79RR101 pKa = 11.84RR102 pKa = 11.84PNNLPGFRR110 pKa = 11.84PLRR113 pKa = 11.84PGPRR117 pKa = 11.84TGNVPRR123 pKa = 11.84MRR125 pKa = 11.84RR126 pKa = 11.84RR127 pKa = 11.84HH128 pKa = 5.89APPSQPSRR136 pKa = 11.84PRR138 pKa = 11.84RR139 pKa = 11.84IAPRR143 pKa = 11.84TTQRR147 pKa = 11.84RR148 pKa = 11.84QSPRR152 pKa = 11.84SRR154 pKa = 11.84GPPRR158 pKa = 11.84SRR160 pKa = 11.84PTPRR164 pKa = 11.84HH165 pKa = 5.64PPRR168 pKa = 11.84SRR170 pKa = 11.84SPRR173 pKa = 11.84PLPTT177 pKa = 3.53
Molecular weight: 20.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
233055 |
44 |
2322 |
337.3 |
37.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.038 ± 0.131 | 1.719 ± 0.044 |
5.326 ± 0.07 | 6.202 ± 0.095 |
4.339 ± 0.069 | 6.341 ± 0.094 |
2.28 ± 0.046 | 5.766 ± 0.074 |
5.306 ± 0.099 | 9.507 ± 0.089 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.294 ± 0.044 | 3.996 ± 0.069 |
4.836 ± 0.097 | 3.613 ± 0.055 |
5.752 ± 0.098 | 8.73 ± 0.145 |
5.408 ± 0.073 | 6.883 ± 0.076 |
1.137 ± 0.032 | 2.524 ± 0.048 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
