
Cryptococcus amylolentus CBS 6039
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Tremellomycetes; Tremellales; Cryptococcaceae; Cryptococcus; Cryptococcus amylolentus
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
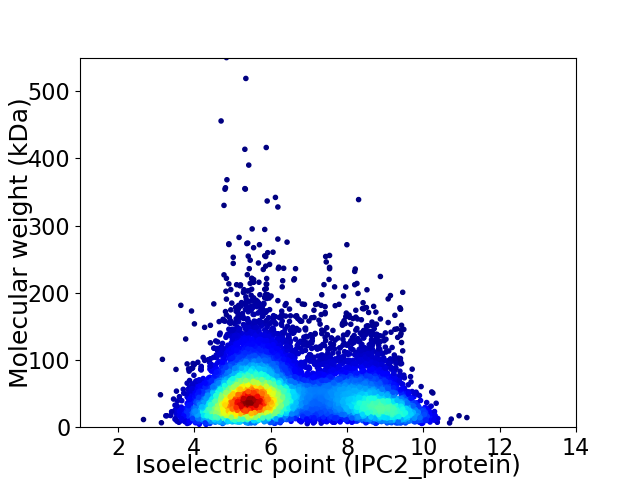
Virtual 2D-PAGE plot for 9877 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
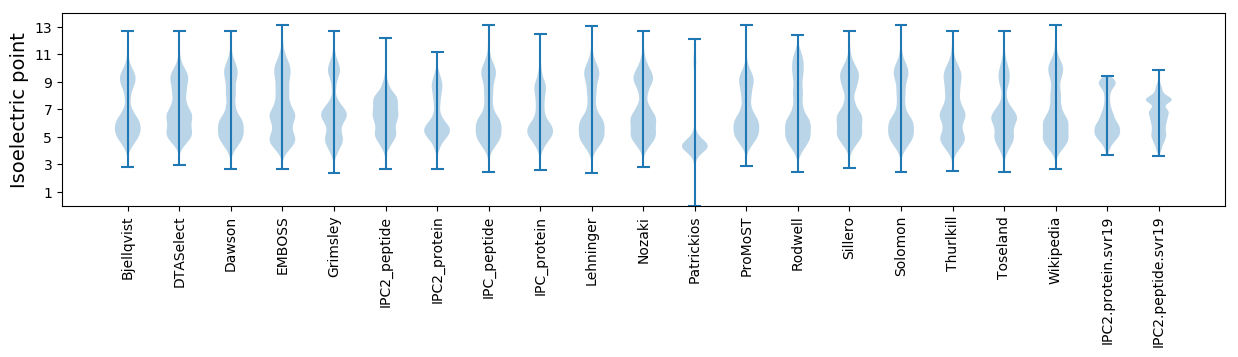
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E3H8V4|A0A1E3H8V4_9TREE Uncharacterized protein OS=Cryptococcus amylolentus CBS 6039 OX=1295533 GN=L202_08203 PE=4 SV=1
MM1 pKa = 7.46RR2 pKa = 11.84QGLFLSILPFLARR15 pKa = 11.84GVLANTCKK23 pKa = 10.56ARR25 pKa = 11.84GASSTSAADD34 pKa = 3.41AADD37 pKa = 3.94AASVDD42 pKa = 3.83GDD44 pKa = 3.92ASSSAADD51 pKa = 3.03IGGGLFASGGASTSGDD67 pKa = 2.98AAASVTSAEE76 pKa = 4.13ASVVTSAGQTAASGGASGEE95 pKa = 4.24AGDD98 pKa = 4.25TDD100 pKa = 3.32IAAAATSATLALTLAASDD118 pKa = 3.89AANATSTVSSAATRR132 pKa = 11.84GSASASSSATSSADD146 pKa = 3.39DD147 pKa = 3.76PMGTQVGTDD156 pKa = 3.62LVLGEE161 pKa = 4.53ASEE164 pKa = 4.46DD165 pKa = 3.86CKK167 pKa = 10.89CGYY170 pKa = 9.84KK171 pKa = 9.95ISSLNDD177 pKa = 2.86QYY179 pKa = 11.61FPYY182 pKa = 10.29QFSFAFSSVDD192 pKa = 3.59DD193 pKa = 4.66GSADD197 pKa = 3.83SLTDD201 pKa = 3.56QKK203 pKa = 11.07WVLNDD208 pKa = 3.06GDD210 pKa = 4.54YY211 pKa = 11.55VGGAANGNRR220 pKa = 11.84CYY222 pKa = 10.82GKK224 pKa = 10.43KK225 pKa = 10.33DD226 pKa = 3.32NLYY229 pKa = 9.68IDD231 pKa = 4.42GGNLVLKK238 pKa = 10.31VPKK241 pKa = 10.33DD242 pKa = 3.47QTASPNMEE250 pKa = 4.08CAEE253 pKa = 4.05IAFEE257 pKa = 4.22EE258 pKa = 4.59TNITGGIFQTTAMLSGVDD276 pKa = 3.98GTCQAFWLNHH286 pKa = 6.22SIATQYY292 pKa = 11.29ADD294 pKa = 3.45EE295 pKa = 4.37VDD297 pKa = 3.47IEE299 pKa = 4.8VISSTIDD306 pKa = 3.1TDD308 pKa = 4.93GIWYY312 pKa = 10.04SNWPPNGDD320 pKa = 3.89PNDD323 pKa = 4.12PDD325 pKa = 3.88NLVSAHH331 pKa = 5.96TNVAVPDD338 pKa = 3.99IDD340 pKa = 4.67SSDD343 pKa = 3.4PRR345 pKa = 11.84KK346 pKa = 8.58TYY348 pKa = 11.29NNYY351 pKa = 8.89TIAWLDD357 pKa = 3.54DD358 pKa = 3.44STNRR362 pKa = 11.84YY363 pKa = 9.31YY364 pKa = 11.21NGAKK368 pKa = 9.45QDD370 pKa = 3.97SPTEE374 pKa = 4.02NQPEE378 pKa = 4.09HH379 pKa = 8.82SMMFVINNWSNGGSGWTLGPPTGEE403 pKa = 4.4DD404 pKa = 2.87SLLKK408 pKa = 10.5VKK410 pKa = 10.62SVLLYY415 pKa = 10.9YY416 pKa = 9.43KK417 pKa = 9.41TANSTQMSDD426 pKa = 4.27LGGDD430 pKa = 3.74CQEE433 pKa = 4.88SDD435 pKa = 3.76VCTVV439 pKa = 2.84
MM1 pKa = 7.46RR2 pKa = 11.84QGLFLSILPFLARR15 pKa = 11.84GVLANTCKK23 pKa = 10.56ARR25 pKa = 11.84GASSTSAADD34 pKa = 3.41AADD37 pKa = 3.94AASVDD42 pKa = 3.83GDD44 pKa = 3.92ASSSAADD51 pKa = 3.03IGGGLFASGGASTSGDD67 pKa = 2.98AAASVTSAEE76 pKa = 4.13ASVVTSAGQTAASGGASGEE95 pKa = 4.24AGDD98 pKa = 4.25TDD100 pKa = 3.32IAAAATSATLALTLAASDD118 pKa = 3.89AANATSTVSSAATRR132 pKa = 11.84GSASASSSATSSADD146 pKa = 3.39DD147 pKa = 3.76PMGTQVGTDD156 pKa = 3.62LVLGEE161 pKa = 4.53ASEE164 pKa = 4.46DD165 pKa = 3.86CKK167 pKa = 10.89CGYY170 pKa = 9.84KK171 pKa = 9.95ISSLNDD177 pKa = 2.86QYY179 pKa = 11.61FPYY182 pKa = 10.29QFSFAFSSVDD192 pKa = 3.59DD193 pKa = 4.66GSADD197 pKa = 3.83SLTDD201 pKa = 3.56QKK203 pKa = 11.07WVLNDD208 pKa = 3.06GDD210 pKa = 4.54YY211 pKa = 11.55VGGAANGNRR220 pKa = 11.84CYY222 pKa = 10.82GKK224 pKa = 10.43KK225 pKa = 10.33DD226 pKa = 3.32NLYY229 pKa = 9.68IDD231 pKa = 4.42GGNLVLKK238 pKa = 10.31VPKK241 pKa = 10.33DD242 pKa = 3.47QTASPNMEE250 pKa = 4.08CAEE253 pKa = 4.05IAFEE257 pKa = 4.22EE258 pKa = 4.59TNITGGIFQTTAMLSGVDD276 pKa = 3.98GTCQAFWLNHH286 pKa = 6.22SIATQYY292 pKa = 11.29ADD294 pKa = 3.45EE295 pKa = 4.37VDD297 pKa = 3.47IEE299 pKa = 4.8VISSTIDD306 pKa = 3.1TDD308 pKa = 4.93GIWYY312 pKa = 10.04SNWPPNGDD320 pKa = 3.89PNDD323 pKa = 4.12PDD325 pKa = 3.88NLVSAHH331 pKa = 5.96TNVAVPDD338 pKa = 3.99IDD340 pKa = 4.67SSDD343 pKa = 3.4PRR345 pKa = 11.84KK346 pKa = 8.58TYY348 pKa = 11.29NNYY351 pKa = 8.89TIAWLDD357 pKa = 3.54DD358 pKa = 3.44STNRR362 pKa = 11.84YY363 pKa = 9.31YY364 pKa = 11.21NGAKK368 pKa = 9.45QDD370 pKa = 3.97SPTEE374 pKa = 4.02NQPEE378 pKa = 4.09HH379 pKa = 8.82SMMFVINNWSNGGSGWTLGPPTGEE403 pKa = 4.4DD404 pKa = 2.87SLLKK408 pKa = 10.5VKK410 pKa = 10.62SVLLYY415 pKa = 10.9YY416 pKa = 9.43KK417 pKa = 9.41TANSTQMSDD426 pKa = 4.27LGGDD430 pKa = 3.74CQEE433 pKa = 4.88SDD435 pKa = 3.76VCTVV439 pKa = 2.84
Molecular weight: 45.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E3HCD1|A0A1E3HCD1_9TREE Uncharacterized protein OS=Cryptococcus amylolentus CBS 6039 OX=1295533 GN=L202_07487 PE=3 SV=1
MM1 pKa = 7.64SFLRR5 pKa = 11.84LNILRR10 pKa = 11.84PALSSSTPLPRR21 pKa = 11.84AQAFSSSSASSRR33 pKa = 11.84PSLLSLSPLSSASSSPLASLRR54 pKa = 11.84FRR56 pKa = 11.84SRR58 pKa = 11.84QQRR61 pKa = 11.84LPLGPGAVGGMGLGGSGRR79 pKa = 11.84RR80 pKa = 11.84EE81 pKa = 4.01FGWTRR86 pKa = 11.84VVEE89 pKa = 4.32ANPQKK94 pKa = 8.61QTKK97 pKa = 9.0LKK99 pKa = 10.09SHH101 pKa = 6.43SSSKK105 pKa = 10.62KK106 pKa = 9.97RR107 pKa = 11.84FFANAKK113 pKa = 10.08GMFKK117 pKa = 10.42RR118 pKa = 11.84AQAGKK123 pKa = 9.97AHH125 pKa = 7.18LNTPLSSSSINRR137 pKa = 11.84LAKK140 pKa = 9.93GVRR143 pKa = 11.84VTNTQARR150 pKa = 11.84KK151 pKa = 9.28LRR153 pKa = 11.84KK154 pKa = 9.17LLPFAA159 pKa = 5.4
MM1 pKa = 7.64SFLRR5 pKa = 11.84LNILRR10 pKa = 11.84PALSSSTPLPRR21 pKa = 11.84AQAFSSSSASSRR33 pKa = 11.84PSLLSLSPLSSASSSPLASLRR54 pKa = 11.84FRR56 pKa = 11.84SRR58 pKa = 11.84QQRR61 pKa = 11.84LPLGPGAVGGMGLGGSGRR79 pKa = 11.84RR80 pKa = 11.84EE81 pKa = 4.01FGWTRR86 pKa = 11.84VVEE89 pKa = 4.32ANPQKK94 pKa = 8.61QTKK97 pKa = 9.0LKK99 pKa = 10.09SHH101 pKa = 6.43SSSKK105 pKa = 10.62KK106 pKa = 9.97RR107 pKa = 11.84FFANAKK113 pKa = 10.08GMFKK117 pKa = 10.42RR118 pKa = 11.84AQAGKK123 pKa = 9.97AHH125 pKa = 7.18LNTPLSSSSINRR137 pKa = 11.84LAKK140 pKa = 9.93GVRR143 pKa = 11.84VTNTQARR150 pKa = 11.84KK151 pKa = 9.28LRR153 pKa = 11.84KK154 pKa = 9.17LLPFAA159 pKa = 5.4
Molecular weight: 17.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4861164 |
41 |
4967 |
492.2 |
53.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.823 ± 0.022 | 1.008 ± 0.008 |
5.485 ± 0.019 | 6.508 ± 0.023 |
3.363 ± 0.016 | 7.406 ± 0.024 |
2.353 ± 0.011 | 4.444 ± 0.017 |
4.97 ± 0.022 | 8.872 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.09 ± 0.008 | 3.12 ± 0.012 |
6.925 ± 0.029 | 3.813 ± 0.018 |
5.95 ± 0.022 | 9.012 ± 0.033 |
5.764 ± 0.016 | 6.131 ± 0.017 |
1.413 ± 0.009 | 2.551 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
