
Arboricoccus pini
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Geminicoccaceae; Arboricoccus
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
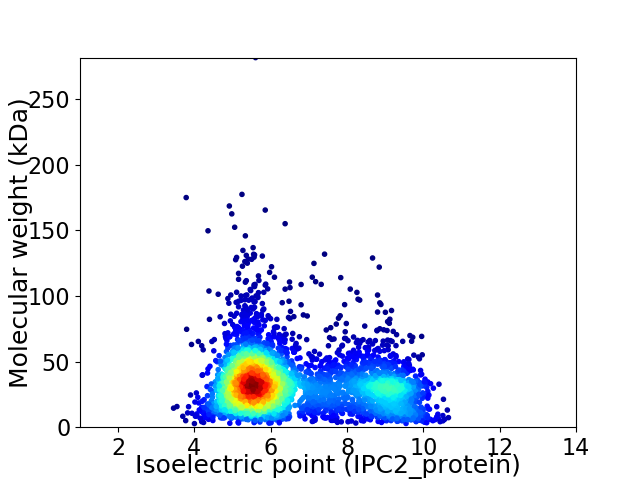
Virtual 2D-PAGE plot for 3717 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A212RWQ3|A0A212RWQ3_9PROT DNA-binding transcriptional regulator LysR family OS=Arboricoccus pini OX=1963835 GN=SAMN07250955_11638 PE=3 SV=1
MM1 pKa = 7.31SADD4 pKa = 3.17SATRR8 pKa = 11.84ARR10 pKa = 11.84DD11 pKa = 3.45MIDD14 pKa = 3.33NIGINIHH21 pKa = 6.79LSYY24 pKa = 11.18TDD26 pKa = 3.93GDD28 pKa = 4.01YY29 pKa = 11.46KK30 pKa = 11.34NYY32 pKa = 10.75DD33 pKa = 3.42SVIADD38 pKa = 4.43LGYY41 pKa = 11.09LGITLVRR48 pKa = 11.84DD49 pKa = 3.65RR50 pKa = 11.84VPNPNTNGGGGKK62 pKa = 10.06NYY64 pKa = 10.61GYY66 pKa = 10.53LADD69 pKa = 4.04AGIRR73 pKa = 11.84FDD75 pKa = 5.1LNASAAASDD84 pKa = 4.13FAGTMTLIEE93 pKa = 4.6NFVAAHH99 pKa = 6.66PGSVVAIEE107 pKa = 4.47GANEE111 pKa = 3.68VDD113 pKa = 4.33KK114 pKa = 11.81NPVSYY119 pKa = 11.0DD120 pKa = 3.13GLTGQAAALALQSDD134 pKa = 4.4IYY136 pKa = 11.11DD137 pKa = 3.75YY138 pKa = 11.83VHH140 pKa = 7.0TSTTLEE146 pKa = 5.38GIATWDD152 pKa = 3.28LTGITGGTDD161 pKa = 3.26FADD164 pKa = 3.56ASNAHH169 pKa = 7.0IYY171 pKa = 8.77PRR173 pKa = 11.84NGNQPQDD180 pKa = 3.46RR181 pKa = 11.84LGLAIEE187 pKa = 4.48TTLAAQPDD195 pKa = 4.11RR196 pKa = 11.84PMVITEE202 pKa = 4.31GGYY205 pKa = 8.45YY206 pKa = 9.38TLPGQGWGGVDD217 pKa = 3.12ATTQSIYY224 pKa = 10.19TINYY228 pKa = 8.95LLDD231 pKa = 3.79AVSQGVSTTFLYY243 pKa = 10.68QLLDD247 pKa = 3.81AYY249 pKa = 9.92TDD251 pKa = 3.52TTGTDD256 pKa = 3.12IEE258 pKa = 4.47KK259 pKa = 10.74HH260 pKa = 5.8FGLFDD265 pKa = 4.64ADD267 pKa = 4.0NNPKK271 pKa = 9.22QAAVALHH278 pKa = 6.43NFTTILSDD286 pKa = 3.53TGASATTFAVDD297 pKa = 3.17TLDD300 pKa = 4.0YY301 pKa = 11.3DD302 pKa = 3.91VTGWNSYY309 pKa = 7.3GHH311 pKa = 6.54SYY313 pKa = 10.95LFEE316 pKa = 6.17KK317 pKa = 10.79SDD319 pKa = 3.5GTFDD323 pKa = 2.99IALWYY328 pKa = 9.36EE329 pKa = 3.52QDD331 pKa = 3.04IWDD334 pKa = 4.37EE335 pKa = 4.39VNLVPIATTSTAIDD349 pKa = 3.96LALGSYY355 pKa = 9.79FRR357 pKa = 11.84VVEE360 pKa = 4.15VFDD363 pKa = 4.21PQLGTGAIATYY374 pKa = 10.64HH375 pKa = 6.86DD376 pKa = 4.55VDD378 pKa = 4.76HH379 pKa = 7.4LDD381 pKa = 3.7LLVNQDD387 pKa = 3.48ALIVQLEE394 pKa = 4.28PNGANAAPTIEE405 pKa = 4.32GVNPQQVVVGDD416 pKa = 4.35DD417 pKa = 3.51LTLSFTSDD425 pKa = 3.58MISDD429 pKa = 4.05ANGDD433 pKa = 4.03TVLLNATLANGSALPSWLTFDD454 pKa = 4.58PDD456 pKa = 3.0TWTLSGVATQTGKK469 pKa = 10.13WSILLTASDD478 pKa = 4.42HH479 pKa = 6.21YY480 pKa = 11.32GGASSLVFQLSVTSQDD496 pKa = 3.54HH497 pKa = 5.76EE498 pKa = 4.26QTPTITGDD506 pKa = 3.22GTIIGTKK513 pKa = 9.77AADD516 pKa = 3.82VIAGGDD522 pKa = 3.42NSDD525 pKa = 4.53TINASSGADD534 pKa = 2.72IVYY537 pKa = 10.05GHH539 pKa = 7.57AGNDD543 pKa = 4.05LINGGGDD550 pKa = 3.47ADD552 pKa = 4.41DD553 pKa = 5.34LYY555 pKa = 11.64GGAGNDD561 pKa = 3.79TLRR564 pKa = 11.84GDD566 pKa = 3.77AGADD570 pKa = 3.19RR571 pKa = 11.84LFGQGGDD578 pKa = 3.59DD579 pKa = 3.57TLAGGGGGFDD589 pKa = 5.04LLTGGAGADD598 pKa = 3.3TFIFRR603 pKa = 11.84GKK605 pKa = 10.23EE606 pKa = 3.5STGPVEE612 pKa = 4.21SVEE615 pKa = 4.1GFLVVQHH622 pKa = 6.41ARR624 pKa = 11.84IADD627 pKa = 3.67LDD629 pKa = 3.84FAEE632 pKa = 4.92GDD634 pKa = 3.8RR635 pKa = 11.84LQLSYY640 pKa = 10.7FVDD643 pKa = 3.49SSGVSLQKK651 pKa = 10.89AVGLSANIDD660 pKa = 3.83SLSGLGTLVHH670 pKa = 6.87FLEE673 pKa = 5.04TDD675 pKa = 3.57DD676 pKa = 4.68PSHH679 pKa = 6.85VIADD683 pKa = 4.05ASTDD687 pKa = 3.52TLTLILKK694 pKa = 9.94DD695 pKa = 3.9SNGGLHH701 pKa = 6.92ALEE704 pKa = 4.61LLHH707 pKa = 6.46YY708 pKa = 9.25ATIAA712 pKa = 3.26
MM1 pKa = 7.31SADD4 pKa = 3.17SATRR8 pKa = 11.84ARR10 pKa = 11.84DD11 pKa = 3.45MIDD14 pKa = 3.33NIGINIHH21 pKa = 6.79LSYY24 pKa = 11.18TDD26 pKa = 3.93GDD28 pKa = 4.01YY29 pKa = 11.46KK30 pKa = 11.34NYY32 pKa = 10.75DD33 pKa = 3.42SVIADD38 pKa = 4.43LGYY41 pKa = 11.09LGITLVRR48 pKa = 11.84DD49 pKa = 3.65RR50 pKa = 11.84VPNPNTNGGGGKK62 pKa = 10.06NYY64 pKa = 10.61GYY66 pKa = 10.53LADD69 pKa = 4.04AGIRR73 pKa = 11.84FDD75 pKa = 5.1LNASAAASDD84 pKa = 4.13FAGTMTLIEE93 pKa = 4.6NFVAAHH99 pKa = 6.66PGSVVAIEE107 pKa = 4.47GANEE111 pKa = 3.68VDD113 pKa = 4.33KK114 pKa = 11.81NPVSYY119 pKa = 11.0DD120 pKa = 3.13GLTGQAAALALQSDD134 pKa = 4.4IYY136 pKa = 11.11DD137 pKa = 3.75YY138 pKa = 11.83VHH140 pKa = 7.0TSTTLEE146 pKa = 5.38GIATWDD152 pKa = 3.28LTGITGGTDD161 pKa = 3.26FADD164 pKa = 3.56ASNAHH169 pKa = 7.0IYY171 pKa = 8.77PRR173 pKa = 11.84NGNQPQDD180 pKa = 3.46RR181 pKa = 11.84LGLAIEE187 pKa = 4.48TTLAAQPDD195 pKa = 4.11RR196 pKa = 11.84PMVITEE202 pKa = 4.31GGYY205 pKa = 8.45YY206 pKa = 9.38TLPGQGWGGVDD217 pKa = 3.12ATTQSIYY224 pKa = 10.19TINYY228 pKa = 8.95LLDD231 pKa = 3.79AVSQGVSTTFLYY243 pKa = 10.68QLLDD247 pKa = 3.81AYY249 pKa = 9.92TDD251 pKa = 3.52TTGTDD256 pKa = 3.12IEE258 pKa = 4.47KK259 pKa = 10.74HH260 pKa = 5.8FGLFDD265 pKa = 4.64ADD267 pKa = 4.0NNPKK271 pKa = 9.22QAAVALHH278 pKa = 6.43NFTTILSDD286 pKa = 3.53TGASATTFAVDD297 pKa = 3.17TLDD300 pKa = 4.0YY301 pKa = 11.3DD302 pKa = 3.91VTGWNSYY309 pKa = 7.3GHH311 pKa = 6.54SYY313 pKa = 10.95LFEE316 pKa = 6.17KK317 pKa = 10.79SDD319 pKa = 3.5GTFDD323 pKa = 2.99IALWYY328 pKa = 9.36EE329 pKa = 3.52QDD331 pKa = 3.04IWDD334 pKa = 4.37EE335 pKa = 4.39VNLVPIATTSTAIDD349 pKa = 3.96LALGSYY355 pKa = 9.79FRR357 pKa = 11.84VVEE360 pKa = 4.15VFDD363 pKa = 4.21PQLGTGAIATYY374 pKa = 10.64HH375 pKa = 6.86DD376 pKa = 4.55VDD378 pKa = 4.76HH379 pKa = 7.4LDD381 pKa = 3.7LLVNQDD387 pKa = 3.48ALIVQLEE394 pKa = 4.28PNGANAAPTIEE405 pKa = 4.32GVNPQQVVVGDD416 pKa = 4.35DD417 pKa = 3.51LTLSFTSDD425 pKa = 3.58MISDD429 pKa = 4.05ANGDD433 pKa = 4.03TVLLNATLANGSALPSWLTFDD454 pKa = 4.58PDD456 pKa = 3.0TWTLSGVATQTGKK469 pKa = 10.13WSILLTASDD478 pKa = 4.42HH479 pKa = 6.21YY480 pKa = 11.32GGASSLVFQLSVTSQDD496 pKa = 3.54HH497 pKa = 5.76EE498 pKa = 4.26QTPTITGDD506 pKa = 3.22GTIIGTKK513 pKa = 9.77AADD516 pKa = 3.82VIAGGDD522 pKa = 3.42NSDD525 pKa = 4.53TINASSGADD534 pKa = 2.72IVYY537 pKa = 10.05GHH539 pKa = 7.57AGNDD543 pKa = 4.05LINGGGDD550 pKa = 3.47ADD552 pKa = 4.41DD553 pKa = 5.34LYY555 pKa = 11.64GGAGNDD561 pKa = 3.79TLRR564 pKa = 11.84GDD566 pKa = 3.77AGADD570 pKa = 3.19RR571 pKa = 11.84LFGQGGDD578 pKa = 3.59DD579 pKa = 3.57TLAGGGGGFDD589 pKa = 5.04LLTGGAGADD598 pKa = 3.3TFIFRR603 pKa = 11.84GKK605 pKa = 10.23EE606 pKa = 3.5STGPVEE612 pKa = 4.21SVEE615 pKa = 4.1GFLVVQHH622 pKa = 6.41ARR624 pKa = 11.84IADD627 pKa = 3.67LDD629 pKa = 3.84FAEE632 pKa = 4.92GDD634 pKa = 3.8RR635 pKa = 11.84LQLSYY640 pKa = 10.7FVDD643 pKa = 3.49SSGVSLQKK651 pKa = 10.89AVGLSANIDD660 pKa = 3.83SLSGLGTLVHH670 pKa = 6.87FLEE673 pKa = 5.04TDD675 pKa = 3.57DD676 pKa = 4.68PSHH679 pKa = 6.85VIADD683 pKa = 4.05ASTDD687 pKa = 3.52TLTLILKK694 pKa = 9.94DD695 pKa = 3.9SNGGLHH701 pKa = 6.92ALEE704 pKa = 4.61LLHH707 pKa = 6.46YY708 pKa = 9.25ATIAA712 pKa = 3.26
Molecular weight: 74.6 kDa
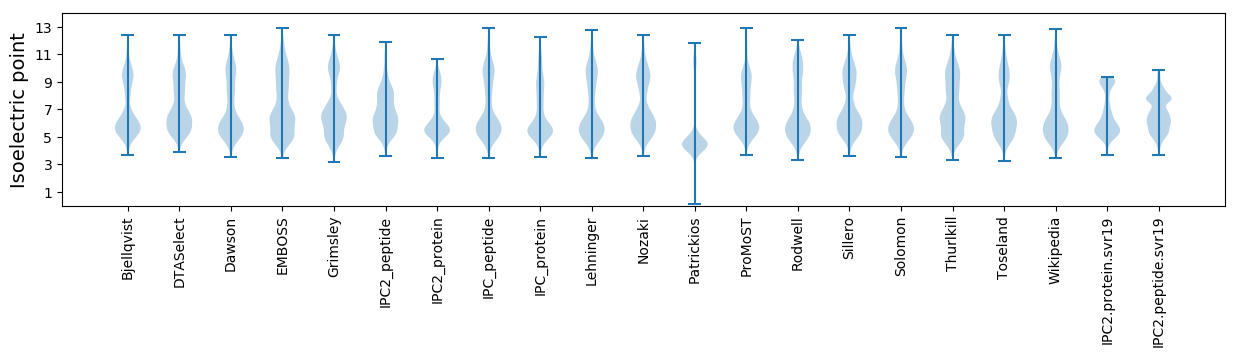
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A212QSZ5|A0A212QSZ5_9PROT Uncharacterized protein OS=Arboricoccus pini OX=1963835 GN=SAMN07250955_103173 PE=4 SV=1
MM1 pKa = 7.44LVLGLTGSLAMGKK14 pKa = 9.57SRR16 pKa = 11.84TAAMFMAHH24 pKa = 7.73GIPVFDD30 pKa = 4.68SDD32 pKa = 3.9ATVHH36 pKa = 6.3GLYY39 pKa = 10.58APGGAAVLPILRR51 pKa = 11.84LFEE54 pKa = 5.02DD55 pKa = 4.53CGSVSDD61 pKa = 5.08GIDD64 pKa = 2.99RR65 pKa = 11.84AALGRR70 pKa = 11.84RR71 pKa = 11.84ILSDD75 pKa = 3.49DD76 pKa = 3.19TAMRR80 pKa = 11.84ALEE83 pKa = 4.22RR84 pKa = 11.84VVHH87 pKa = 5.83ALVRR91 pKa = 11.84SSQRR95 pKa = 11.84AFLEE99 pKa = 4.21TQARR103 pKa = 11.84LGHH106 pKa = 6.81WLTVLDD112 pKa = 4.52IPLLYY117 pKa = 10.54EE118 pKa = 3.92SGGDD122 pKa = 3.5RR123 pKa = 11.84RR124 pKa = 11.84CDD126 pKa = 3.48RR127 pKa = 11.84IVVVSASPFIQSQRR141 pKa = 11.84ALARR145 pKa = 11.84TGMTALRR152 pKa = 11.84LKK154 pKa = 10.71SLLEE158 pKa = 4.03RR159 pKa = 11.84QTPDD163 pKa = 4.14LRR165 pKa = 11.84KK166 pKa = 9.37QCLADD171 pKa = 4.1FVIHH175 pKa = 6.38TGGDD179 pKa = 3.07RR180 pKa = 11.84GRR182 pKa = 11.84LSRR185 pKa = 11.84NVAEE189 pKa = 5.18ILRR192 pKa = 11.84KK193 pKa = 8.48MRR195 pKa = 11.84QVQPHH200 pKa = 6.84AWPKK204 pKa = 9.62RR205 pKa = 11.84WW206 pKa = 3.14
MM1 pKa = 7.44LVLGLTGSLAMGKK14 pKa = 9.57SRR16 pKa = 11.84TAAMFMAHH24 pKa = 7.73GIPVFDD30 pKa = 4.68SDD32 pKa = 3.9ATVHH36 pKa = 6.3GLYY39 pKa = 10.58APGGAAVLPILRR51 pKa = 11.84LFEE54 pKa = 5.02DD55 pKa = 4.53CGSVSDD61 pKa = 5.08GIDD64 pKa = 2.99RR65 pKa = 11.84AALGRR70 pKa = 11.84RR71 pKa = 11.84ILSDD75 pKa = 3.49DD76 pKa = 3.19TAMRR80 pKa = 11.84ALEE83 pKa = 4.22RR84 pKa = 11.84VVHH87 pKa = 5.83ALVRR91 pKa = 11.84SSQRR95 pKa = 11.84AFLEE99 pKa = 4.21TQARR103 pKa = 11.84LGHH106 pKa = 6.81WLTVLDD112 pKa = 4.52IPLLYY117 pKa = 10.54EE118 pKa = 3.92SGGDD122 pKa = 3.5RR123 pKa = 11.84RR124 pKa = 11.84CDD126 pKa = 3.48RR127 pKa = 11.84IVVVSASPFIQSQRR141 pKa = 11.84ALARR145 pKa = 11.84TGMTALRR152 pKa = 11.84LKK154 pKa = 10.71SLLEE158 pKa = 4.03RR159 pKa = 11.84QTPDD163 pKa = 4.14LRR165 pKa = 11.84KK166 pKa = 9.37QCLADD171 pKa = 4.1FVIHH175 pKa = 6.38TGGDD179 pKa = 3.07RR180 pKa = 11.84GRR182 pKa = 11.84LSRR185 pKa = 11.84NVAEE189 pKa = 5.18ILRR192 pKa = 11.84KK193 pKa = 8.48MRR195 pKa = 11.84QVQPHH200 pKa = 6.84AWPKK204 pKa = 9.62RR205 pKa = 11.84WW206 pKa = 3.14
Molecular weight: 22.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1199682 |
24 |
2625 |
322.8 |
35.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.565 ± 0.054 | 0.861 ± 0.013 |
5.639 ± 0.029 | 5.532 ± 0.036 |
3.578 ± 0.025 | 8.572 ± 0.04 |
2.083 ± 0.018 | 5.168 ± 0.032 |
2.912 ± 0.03 | 11.092 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.435 ± 0.017 | 2.321 ± 0.021 |
5.419 ± 0.028 | 3.263 ± 0.025 |
7.524 ± 0.043 | 5.307 ± 0.031 |
5.101 ± 0.028 | 7.168 ± 0.03 |
1.32 ± 0.015 | 2.14 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
