
Arthrobacter phage Judy
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Bridgettevirus; Arthrobacter virus Judy
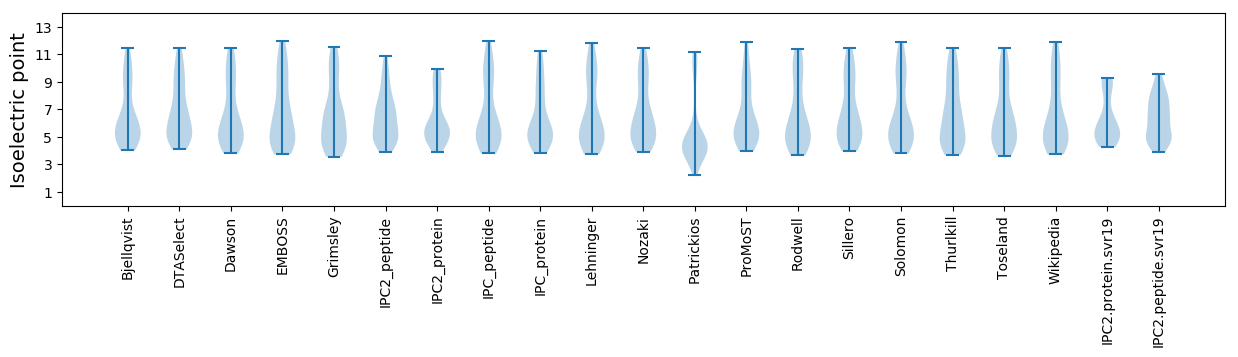
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 72 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A3G2KGP0|A0A3G2KGP0_9CAUD Uncharacterized protein OS=Arthrobacter phage Judy OX=2419958 GN=65 PE=4 SV=1
MM1 pKa = 7.06NHH3 pKa = 5.9NFKK6 pKa = 10.31EE7 pKa = 4.66VPSTEE12 pKa = 4.19TPDD15 pKa = 5.47DD16 pKa = 4.26GDD18 pKa = 3.41LTLALRR24 pKa = 11.84QRR26 pKa = 11.84AHH28 pKa = 6.04GTLTQQVFGLEE39 pKa = 4.13PYY41 pKa = 10.32IIIRR45 pKa = 11.84PTFAEE50 pKa = 4.12DD51 pKa = 2.73TGAFQVGIEE60 pKa = 4.23TGGGADD66 pKa = 3.69LANVGEE72 pKa = 4.25FLEE75 pKa = 5.32LIAEE79 pKa = 4.78AMQADD84 pKa = 3.84ATKK87 pKa = 10.63EE88 pKa = 3.9NIAAATSGDD97 pKa = 4.31DD98 pKa = 3.79PDD100 pKa = 4.02EE101 pKa = 5.92DD102 pKa = 3.87EE103 pKa = 4.6
MM1 pKa = 7.06NHH3 pKa = 5.9NFKK6 pKa = 10.31EE7 pKa = 4.66VPSTEE12 pKa = 4.19TPDD15 pKa = 5.47DD16 pKa = 4.26GDD18 pKa = 3.41LTLALRR24 pKa = 11.84QRR26 pKa = 11.84AHH28 pKa = 6.04GTLTQQVFGLEE39 pKa = 4.13PYY41 pKa = 10.32IIIRR45 pKa = 11.84PTFAEE50 pKa = 4.12DD51 pKa = 2.73TGAFQVGIEE60 pKa = 4.23TGGGADD66 pKa = 3.69LANVGEE72 pKa = 4.25FLEE75 pKa = 5.32LIAEE79 pKa = 4.78AMQADD84 pKa = 3.84ATKK87 pKa = 10.63EE88 pKa = 3.9NIAAATSGDD97 pKa = 4.31DD98 pKa = 3.79PDD100 pKa = 4.02EE101 pKa = 5.92DD102 pKa = 3.87EE103 pKa = 4.6
Molecular weight: 10.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G2KGG8|A0A3G2KGG8_9CAUD Portal protein OS=Arthrobacter phage Judy OX=2419958 GN=3 PE=4 SV=1
MM1 pKa = 7.14YY2 pKa = 9.47PDD4 pKa = 4.58AATAHH9 pKa = 5.55YY10 pKa = 10.87RR11 pKa = 11.84LMQRR15 pKa = 11.84LQTVAVVAGRR25 pKa = 11.84RR26 pKa = 11.84AWSRR30 pKa = 11.84IDD32 pKa = 3.73PGDD35 pKa = 4.0LSGSWMQGIAVVAGIVTTQQEE56 pKa = 4.13RR57 pKa = 11.84AAVAGAGYY65 pKa = 8.22VTDD68 pKa = 4.66ALAAQGEE75 pKa = 4.85YY76 pKa = 10.11VAPEE80 pKa = 4.09AFLNPAAFAGTAADD94 pKa = 3.89GRR96 pKa = 11.84SLSTLLYY103 pKa = 10.91SPVTTVKK110 pKa = 10.37QQLAEE115 pKa = 4.25GASMASALTAGRR127 pKa = 11.84GALDD131 pKa = 3.12MLLRR135 pKa = 11.84VTVADD140 pKa = 3.91AGRR143 pKa = 11.84VASGVNVATRR153 pKa = 11.84QSVGYY158 pKa = 10.63VRR160 pKa = 11.84MLNPPSCPRR169 pKa = 11.84CSVLAGRR176 pKa = 11.84FYY178 pKa = 10.94RR179 pKa = 11.84WNAGFLRR186 pKa = 11.84HH187 pKa = 6.09PKK189 pKa = 9.45CDD191 pKa = 3.72CVHH194 pKa = 5.82VPSKK198 pKa = 11.03GVDD201 pKa = 3.19AAKK204 pKa = 10.62SEE206 pKa = 4.09GLIHH210 pKa = 7.67DD211 pKa = 4.55PYY213 pKa = 11.06EE214 pKa = 4.36YY215 pKa = 10.58FHH217 pKa = 6.93SLSEE221 pKa = 3.98QEE223 pKa = 4.27QNEE226 pKa = 4.75KK227 pKa = 8.4YY228 pKa = 8.81TKK230 pKa = 10.37AGARR234 pKa = 11.84AIRR237 pKa = 11.84DD238 pKa = 3.43GGDD241 pKa = 2.42IFQVVNSQRR250 pKa = 11.84GMSYY254 pKa = 11.34ASISKK259 pKa = 10.33DD260 pKa = 2.84GTRR263 pKa = 11.84RR264 pKa = 11.84GQRR267 pKa = 11.84STSRR271 pKa = 11.84FTTEE275 pKa = 3.17GTTRR279 pKa = 11.84RR280 pKa = 11.84GNFGRR285 pKa = 11.84KK286 pKa = 8.87GRR288 pKa = 11.84LTPEE292 pKa = 5.35AIYY295 pKa = 9.78QQSGSRR301 pKa = 11.84EE302 pKa = 3.65EE303 pKa = 4.1ALRR306 pKa = 11.84LLAANGYY313 pKa = 9.12ILPGGQNPLGSLRR326 pKa = 11.84GQRR329 pKa = 11.84EE330 pKa = 4.09GFGQLGRR337 pKa = 11.84GGTRR341 pKa = 11.84VGARR345 pKa = 11.84EE346 pKa = 3.87AVLEE350 pKa = 3.87ARR352 pKa = 11.84RR353 pKa = 11.84TGVRR357 pKa = 11.84TGARR361 pKa = 11.84ATMTAAEE368 pKa = 4.58LRR370 pKa = 11.84VFDD373 pKa = 5.47ARR375 pKa = 11.84SRR377 pKa = 11.84WDD379 pKa = 3.35QVRR382 pKa = 11.84KK383 pKa = 9.73GVNPFNPKK391 pKa = 10.09RR392 pKa = 11.84PLTPEE397 pKa = 2.95IAARR401 pKa = 11.84VEE403 pKa = 3.8RR404 pKa = 11.84DD405 pKa = 3.2FRR407 pKa = 11.84KK408 pKa = 10.63YY409 pKa = 10.25ILGYY413 pKa = 10.5
MM1 pKa = 7.14YY2 pKa = 9.47PDD4 pKa = 4.58AATAHH9 pKa = 5.55YY10 pKa = 10.87RR11 pKa = 11.84LMQRR15 pKa = 11.84LQTVAVVAGRR25 pKa = 11.84RR26 pKa = 11.84AWSRR30 pKa = 11.84IDD32 pKa = 3.73PGDD35 pKa = 4.0LSGSWMQGIAVVAGIVTTQQEE56 pKa = 4.13RR57 pKa = 11.84AAVAGAGYY65 pKa = 8.22VTDD68 pKa = 4.66ALAAQGEE75 pKa = 4.85YY76 pKa = 10.11VAPEE80 pKa = 4.09AFLNPAAFAGTAADD94 pKa = 3.89GRR96 pKa = 11.84SLSTLLYY103 pKa = 10.91SPVTTVKK110 pKa = 10.37QQLAEE115 pKa = 4.25GASMASALTAGRR127 pKa = 11.84GALDD131 pKa = 3.12MLLRR135 pKa = 11.84VTVADD140 pKa = 3.91AGRR143 pKa = 11.84VASGVNVATRR153 pKa = 11.84QSVGYY158 pKa = 10.63VRR160 pKa = 11.84MLNPPSCPRR169 pKa = 11.84CSVLAGRR176 pKa = 11.84FYY178 pKa = 10.94RR179 pKa = 11.84WNAGFLRR186 pKa = 11.84HH187 pKa = 6.09PKK189 pKa = 9.45CDD191 pKa = 3.72CVHH194 pKa = 5.82VPSKK198 pKa = 11.03GVDD201 pKa = 3.19AAKK204 pKa = 10.62SEE206 pKa = 4.09GLIHH210 pKa = 7.67DD211 pKa = 4.55PYY213 pKa = 11.06EE214 pKa = 4.36YY215 pKa = 10.58FHH217 pKa = 6.93SLSEE221 pKa = 3.98QEE223 pKa = 4.27QNEE226 pKa = 4.75KK227 pKa = 8.4YY228 pKa = 8.81TKK230 pKa = 10.37AGARR234 pKa = 11.84AIRR237 pKa = 11.84DD238 pKa = 3.43GGDD241 pKa = 2.42IFQVVNSQRR250 pKa = 11.84GMSYY254 pKa = 11.34ASISKK259 pKa = 10.33DD260 pKa = 2.84GTRR263 pKa = 11.84RR264 pKa = 11.84GQRR267 pKa = 11.84STSRR271 pKa = 11.84FTTEE275 pKa = 3.17GTTRR279 pKa = 11.84RR280 pKa = 11.84GNFGRR285 pKa = 11.84KK286 pKa = 8.87GRR288 pKa = 11.84LTPEE292 pKa = 5.35AIYY295 pKa = 9.78QQSGSRR301 pKa = 11.84EE302 pKa = 3.65EE303 pKa = 4.1ALRR306 pKa = 11.84LLAANGYY313 pKa = 9.12ILPGGQNPLGSLRR326 pKa = 11.84GQRR329 pKa = 11.84EE330 pKa = 4.09GFGQLGRR337 pKa = 11.84GGTRR341 pKa = 11.84VGARR345 pKa = 11.84EE346 pKa = 3.87AVLEE350 pKa = 3.87ARR352 pKa = 11.84RR353 pKa = 11.84TGVRR357 pKa = 11.84TGARR361 pKa = 11.84ATMTAAEE368 pKa = 4.58LRR370 pKa = 11.84VFDD373 pKa = 5.47ARR375 pKa = 11.84SRR377 pKa = 11.84WDD379 pKa = 3.35QVRR382 pKa = 11.84KK383 pKa = 9.73GVNPFNPKK391 pKa = 10.09RR392 pKa = 11.84PLTPEE397 pKa = 2.95IAARR401 pKa = 11.84VEE403 pKa = 3.8RR404 pKa = 11.84DD405 pKa = 3.2FRR407 pKa = 11.84KK408 pKa = 10.63YY409 pKa = 10.25ILGYY413 pKa = 10.5
Molecular weight: 44.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13953 |
30 |
1298 |
193.8 |
21.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.911 ± 0.57 | 0.953 ± 0.142 |
6.013 ± 0.174 | 5.784 ± 0.307 |
2.559 ± 0.14 | 8.658 ± 0.358 |
1.957 ± 0.226 | 4.307 ± 0.207 |
4.408 ± 0.308 | 7.74 ± 0.322 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.114 ± 0.145 | 3.017 ± 0.19 |
5.87 ± 0.42 | 4.286 ± 0.404 |
6.515 ± 0.414 | 5.569 ± 0.277 |
7.131 ± 0.362 | 6.852 ± 0.33 |
1.82 ± 0.219 | 2.537 ± 0.125 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
