
Roseospira marina
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Roseospira
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
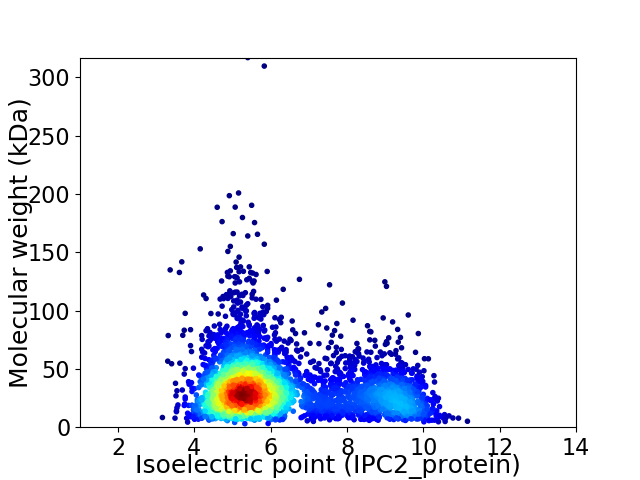
Virtual 2D-PAGE plot for 4009 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
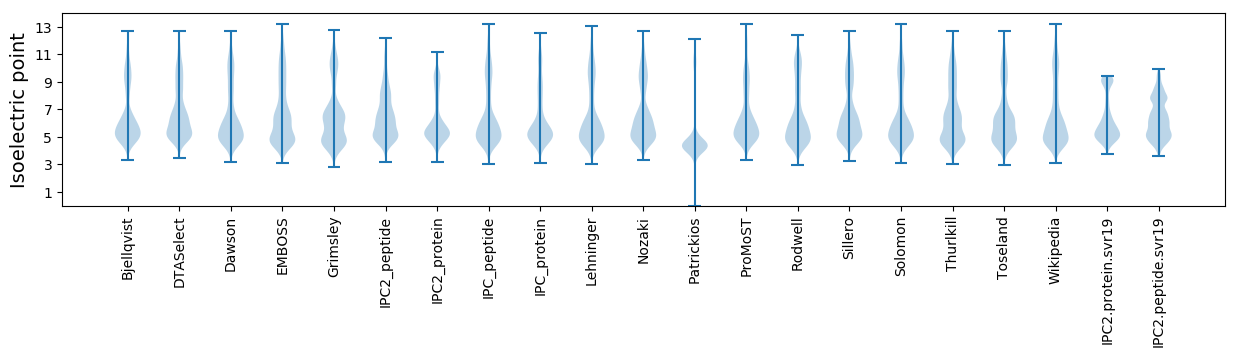
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5M6IAI8|A0A5M6IAI8_9PROT GAF domain-containing protein OS=Roseospira marina OX=140057 GN=F1188_11980 PE=3 SV=1
MM1 pKa = 7.37SVLANALRR9 pKa = 11.84TGTAGAVALALGALGTVGAVAADD32 pKa = 4.54LDD34 pKa = 4.02PGKK37 pKa = 9.84PYY39 pKa = 10.97DD40 pKa = 4.14LGRR43 pKa = 11.84DD44 pKa = 3.58ATPEE48 pKa = 3.89EE49 pKa = 4.29VAGWDD54 pKa = 3.55IDD56 pKa = 3.92VLPDD60 pKa = 3.26GTGALVGQGTPAEE73 pKa = 4.43GEE75 pKa = 4.39AIFMAQCAVCHH86 pKa = 6.2GEE88 pKa = 3.77FGEE91 pKa = 4.74GVDD94 pKa = 4.28RR95 pKa = 11.84YY96 pKa = 9.42PVLFGGEE103 pKa = 4.06DD104 pKa = 3.43TLASRR109 pKa = 11.84DD110 pKa = 3.75PVKK113 pKa = 10.28TPGSYY118 pKa = 8.65WPYY121 pKa = 10.86ASTLVDD127 pKa = 3.6YY128 pKa = 10.17IYY130 pKa = 10.93RR131 pKa = 11.84AMPFGQAQTLTPDD144 pKa = 2.89QAYY147 pKa = 10.61ALTAYY152 pKa = 10.57LLYY155 pKa = 10.51INFVLDD161 pKa = 4.56DD162 pKa = 4.12EE163 pKa = 5.76DD164 pKa = 4.84FVLSNEE170 pKa = 4.37TIGAVEE176 pKa = 4.05MPNRR180 pKa = 11.84DD181 pKa = 2.89GFIRR185 pKa = 11.84DD186 pKa = 4.47DD187 pKa = 4.17RR188 pKa = 11.84PDD190 pKa = 3.67AQPEE194 pKa = 4.57TVCMTACDD202 pKa = 3.76VPTQVVGRR210 pKa = 11.84ARR212 pKa = 11.84IVDD215 pKa = 3.66VTPDD219 pKa = 3.64DD220 pKa = 5.24DD221 pKa = 3.92EE222 pKa = 4.54TAGLSLEE229 pKa = 4.23
MM1 pKa = 7.37SVLANALRR9 pKa = 11.84TGTAGAVALALGALGTVGAVAADD32 pKa = 4.54LDD34 pKa = 4.02PGKK37 pKa = 9.84PYY39 pKa = 10.97DD40 pKa = 4.14LGRR43 pKa = 11.84DD44 pKa = 3.58ATPEE48 pKa = 3.89EE49 pKa = 4.29VAGWDD54 pKa = 3.55IDD56 pKa = 3.92VLPDD60 pKa = 3.26GTGALVGQGTPAEE73 pKa = 4.43GEE75 pKa = 4.39AIFMAQCAVCHH86 pKa = 6.2GEE88 pKa = 3.77FGEE91 pKa = 4.74GVDD94 pKa = 4.28RR95 pKa = 11.84YY96 pKa = 9.42PVLFGGEE103 pKa = 4.06DD104 pKa = 3.43TLASRR109 pKa = 11.84DD110 pKa = 3.75PVKK113 pKa = 10.28TPGSYY118 pKa = 8.65WPYY121 pKa = 10.86ASTLVDD127 pKa = 3.6YY128 pKa = 10.17IYY130 pKa = 10.93RR131 pKa = 11.84AMPFGQAQTLTPDD144 pKa = 2.89QAYY147 pKa = 10.61ALTAYY152 pKa = 10.57LLYY155 pKa = 10.51INFVLDD161 pKa = 4.56DD162 pKa = 4.12EE163 pKa = 5.76DD164 pKa = 4.84FVLSNEE170 pKa = 4.37TIGAVEE176 pKa = 4.05MPNRR180 pKa = 11.84DD181 pKa = 2.89GFIRR185 pKa = 11.84DD186 pKa = 4.47DD187 pKa = 4.17RR188 pKa = 11.84PDD190 pKa = 3.67AQPEE194 pKa = 4.57TVCMTACDD202 pKa = 3.76VPTQVVGRR210 pKa = 11.84ARR212 pKa = 11.84IVDD215 pKa = 3.66VTPDD219 pKa = 3.64DD220 pKa = 5.24DD221 pKa = 3.92EE222 pKa = 4.54TAGLSLEE229 pKa = 4.23
Molecular weight: 24.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5M6IEH4|A0A5M6IEH4_9PROT Tripartite tricarboxylate transporter permease OS=Roseospira marina OX=140057 GN=F1188_04940 PE=4 SV=1
MM1 pKa = 7.39KK2 pKa = 10.27RR3 pKa = 11.84PYY5 pKa = 10.01QPSKK9 pKa = 10.19LVRR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.64GFRR19 pKa = 11.84SRR21 pKa = 11.84SATVGGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.39KK2 pKa = 10.27RR3 pKa = 11.84PYY5 pKa = 10.01QPSKK9 pKa = 10.19LVRR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.64GFRR19 pKa = 11.84SRR21 pKa = 11.84SATVGGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1330878 |
29 |
2901 |
332.0 |
35.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.232 ± 0.064 | 0.89 ± 0.01 |
6.348 ± 0.035 | 5.421 ± 0.03 |
3.163 ± 0.022 | 8.905 ± 0.036 |
2.189 ± 0.019 | 4.293 ± 0.028 |
2.24 ± 0.027 | 10.215 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.445 ± 0.019 | 2.145 ± 0.02 |
5.89 ± 0.034 | 2.784 ± 0.018 |
7.809 ± 0.042 | 4.726 ± 0.023 |
5.985 ± 0.032 | 7.975 ± 0.03 |
1.365 ± 0.016 | 1.978 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
