
Rhizobium sp. Root1212
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium; unclassified Rhizobium
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
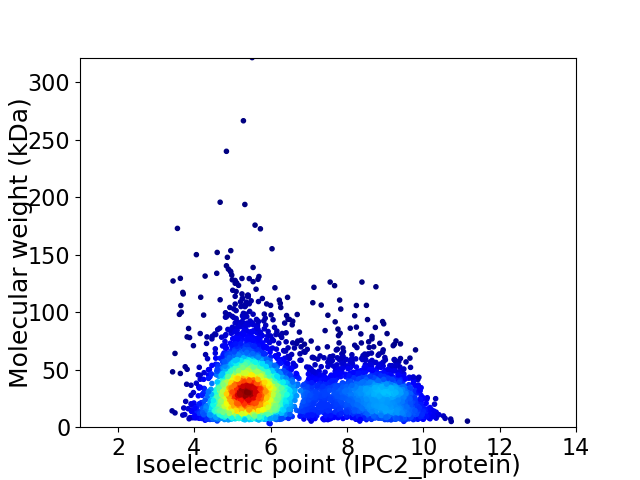
Virtual 2D-PAGE plot for 4824 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q6T3D2|A0A0Q6T3D2_9RHIZ Phosphoglycerate mutase OS=Rhizobium sp. Root1212 OX=1736429 GN=ASC86_05685 PE=4 SV=1
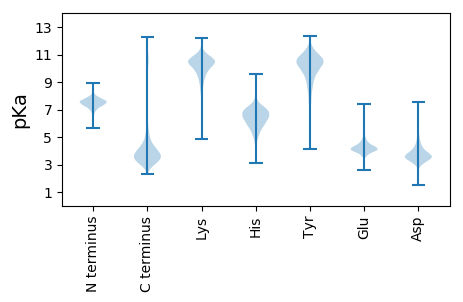
MM1 pKa = 7.49NIKK4 pKa = 10.39SLLLGSAAALAAVSGAHH21 pKa = 6.45AADD24 pKa = 5.02AIVAAEE30 pKa = 4.47PEE32 pKa = 3.93PMEE35 pKa = 4.1YY36 pKa = 10.8VRR38 pKa = 11.84VCDD41 pKa = 4.42AFGTGYY47 pKa = 10.47FYY49 pKa = 10.57IPGTEE54 pKa = 3.93TCLSISGYY62 pKa = 10.28VRR64 pKa = 11.84TQIEE68 pKa = 4.65YY69 pKa = 10.2NDD71 pKa = 3.69SRR73 pKa = 11.84ADD75 pKa = 3.76DD76 pKa = 5.17DD77 pKa = 4.78DD78 pKa = 3.52WGTYY82 pKa = 6.98TRR84 pKa = 11.84AYY86 pKa = 9.6LTFAAKK92 pKa = 10.44NDD94 pKa = 4.01TEE96 pKa = 4.51FGTLSSYY103 pKa = 11.36INLQADD109 pKa = 3.68SGHH112 pKa = 5.71TWDD115 pKa = 4.31VGSGGDD121 pKa = 4.56VILDD125 pKa = 3.79GAWIEE130 pKa = 3.93IAGFKK135 pKa = 9.95IGYY138 pKa = 8.69FYY140 pKa = 11.36NWWDD144 pKa = 3.64DD145 pKa = 3.6FGLAGEE151 pKa = 4.27TDD153 pKa = 3.44EE154 pKa = 5.27TGGNLFNSIQYY165 pKa = 9.47TYY167 pKa = 11.0DD168 pKa = 3.29AGSFQVGASVEE179 pKa = 4.45DD180 pKa = 3.94LDD182 pKa = 6.12ADD184 pKa = 3.82VGSVYY189 pKa = 10.85SADD192 pKa = 3.24QGFGHH197 pKa = 6.74NDD199 pKa = 3.32DD200 pKa = 4.16VGISGLVRR208 pKa = 11.84GTFGGVDD215 pKa = 3.37ALLIASYY222 pKa = 11.26DD223 pKa = 3.63FDD225 pKa = 4.1VEE227 pKa = 4.0EE228 pKa = 4.42AAFKK232 pKa = 10.98ARR234 pKa = 11.84LTAAVGPGTFGIAGIYY250 pKa = 8.57ATDD253 pKa = 3.97PNAYY257 pKa = 8.86WSDD260 pKa = 4.04SEE262 pKa = 4.23WSVVASYY269 pKa = 10.1EE270 pKa = 4.54LKK272 pKa = 10.3ATDD275 pKa = 4.35KK276 pKa = 10.55FTITPAAQYY285 pKa = 9.91FGSLRR290 pKa = 11.84DD291 pKa = 3.76GTNGFGDD298 pKa = 3.72DD299 pKa = 4.31DD300 pKa = 3.63AWKK303 pKa = 10.42VGLTAGYY310 pKa = 10.22QITDD314 pKa = 3.59GLRR317 pKa = 11.84TLATINYY324 pKa = 9.04KK325 pKa = 10.66KK326 pKa = 10.0IDD328 pKa = 3.83TDD330 pKa = 4.14ADD332 pKa = 3.48IDD334 pKa = 4.22DD335 pKa = 4.91DD336 pKa = 4.93SISGFVRR343 pKa = 11.84LQRR346 pKa = 11.84DD347 pKa = 3.62FF348 pKa = 4.22
MM1 pKa = 7.49NIKK4 pKa = 10.39SLLLGSAAALAAVSGAHH21 pKa = 6.45AADD24 pKa = 5.02AIVAAEE30 pKa = 4.47PEE32 pKa = 3.93PMEE35 pKa = 4.1YY36 pKa = 10.8VRR38 pKa = 11.84VCDD41 pKa = 4.42AFGTGYY47 pKa = 10.47FYY49 pKa = 10.57IPGTEE54 pKa = 3.93TCLSISGYY62 pKa = 10.28VRR64 pKa = 11.84TQIEE68 pKa = 4.65YY69 pKa = 10.2NDD71 pKa = 3.69SRR73 pKa = 11.84ADD75 pKa = 3.76DD76 pKa = 5.17DD77 pKa = 4.78DD78 pKa = 3.52WGTYY82 pKa = 6.98TRR84 pKa = 11.84AYY86 pKa = 9.6LTFAAKK92 pKa = 10.44NDD94 pKa = 4.01TEE96 pKa = 4.51FGTLSSYY103 pKa = 11.36INLQADD109 pKa = 3.68SGHH112 pKa = 5.71TWDD115 pKa = 4.31VGSGGDD121 pKa = 4.56VILDD125 pKa = 3.79GAWIEE130 pKa = 3.93IAGFKK135 pKa = 9.95IGYY138 pKa = 8.69FYY140 pKa = 11.36NWWDD144 pKa = 3.64DD145 pKa = 3.6FGLAGEE151 pKa = 4.27TDD153 pKa = 3.44EE154 pKa = 5.27TGGNLFNSIQYY165 pKa = 9.47TYY167 pKa = 11.0DD168 pKa = 3.29AGSFQVGASVEE179 pKa = 4.45DD180 pKa = 3.94LDD182 pKa = 6.12ADD184 pKa = 3.82VGSVYY189 pKa = 10.85SADD192 pKa = 3.24QGFGHH197 pKa = 6.74NDD199 pKa = 3.32DD200 pKa = 4.16VGISGLVRR208 pKa = 11.84GTFGGVDD215 pKa = 3.37ALLIASYY222 pKa = 11.26DD223 pKa = 3.63FDD225 pKa = 4.1VEE227 pKa = 4.0EE228 pKa = 4.42AAFKK232 pKa = 10.98ARR234 pKa = 11.84LTAAVGPGTFGIAGIYY250 pKa = 8.57ATDD253 pKa = 3.97PNAYY257 pKa = 8.86WSDD260 pKa = 4.04SEE262 pKa = 4.23WSVVASYY269 pKa = 10.1EE270 pKa = 4.54LKK272 pKa = 10.3ATDD275 pKa = 4.35KK276 pKa = 10.55FTITPAAQYY285 pKa = 9.91FGSLRR290 pKa = 11.84DD291 pKa = 3.76GTNGFGDD298 pKa = 3.72DD299 pKa = 4.31DD300 pKa = 3.63AWKK303 pKa = 10.42VGLTAGYY310 pKa = 10.22QITDD314 pKa = 3.59GLRR317 pKa = 11.84TLATINYY324 pKa = 9.04KK325 pKa = 10.66KK326 pKa = 10.0IDD328 pKa = 3.83TDD330 pKa = 4.14ADD332 pKa = 3.48IDD334 pKa = 4.22DD335 pKa = 4.91DD336 pKa = 4.93SISGFVRR343 pKa = 11.84LQRR346 pKa = 11.84DD347 pKa = 3.62FF348 pKa = 4.22
Molecular weight: 37.33 kDa
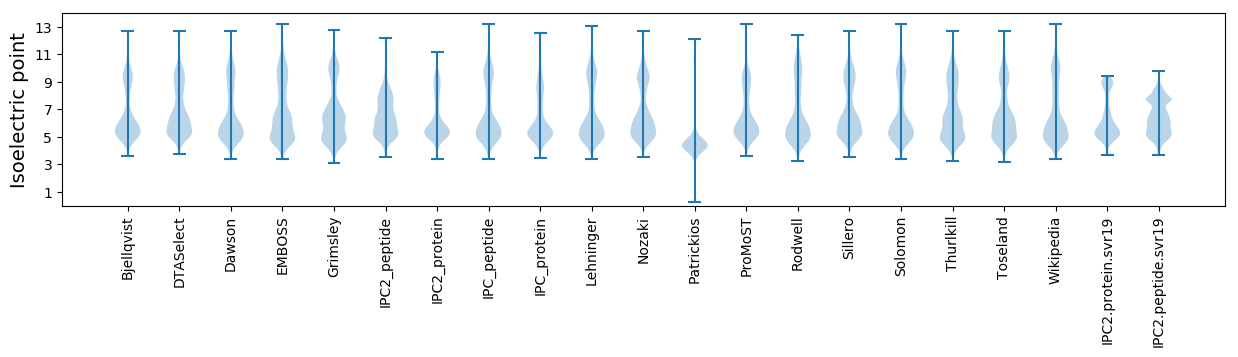
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q6T555|A0A0Q6T555_9RHIZ Uncharacterized protein OS=Rhizobium sp. Root1212 OX=1736429 GN=ASC86_06310 PE=4 SV=1
MM1 pKa = 7.62TKK3 pKa = 9.11RR4 pKa = 11.84TYY6 pKa = 10.36QPSKK10 pKa = 9.73LVRR13 pKa = 11.84KK14 pKa = 9.15RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.64GFRR20 pKa = 11.84ARR22 pKa = 11.84IATKK26 pKa = 9.98GGRR29 pKa = 11.84KK30 pKa = 9.73VIVARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.03RR42 pKa = 11.84LSAA45 pKa = 4.03
MM1 pKa = 7.62TKK3 pKa = 9.11RR4 pKa = 11.84TYY6 pKa = 10.36QPSKK10 pKa = 9.73LVRR13 pKa = 11.84KK14 pKa = 9.15RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.64GFRR20 pKa = 11.84ARR22 pKa = 11.84IATKK26 pKa = 9.98GGRR29 pKa = 11.84KK30 pKa = 9.73VIVARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.03RR42 pKa = 11.84LSAA45 pKa = 4.03
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1524751 |
28 |
2877 |
316.1 |
34.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.913 ± 0.052 | 0.796 ± 0.01 |
5.801 ± 0.026 | 5.817 ± 0.031 |
3.943 ± 0.024 | 8.43 ± 0.036 |
1.976 ± 0.015 | 5.677 ± 0.026 |
3.794 ± 0.026 | 9.951 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.573 ± 0.016 | 2.86 ± 0.023 |
4.804 ± 0.028 | 3.021 ± 0.02 |
6.549 ± 0.037 | 5.734 ± 0.026 |
5.458 ± 0.029 | 7.323 ± 0.028 |
1.239 ± 0.014 | 2.339 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
