
Tortoise microvirus 63
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.35
Get precalculated fractions of proteins
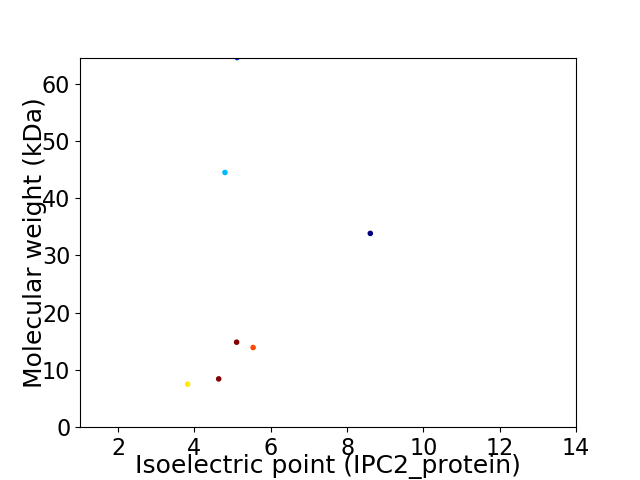
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W7L1|A0A4P8W7L1_9VIRU Uncharacterized protein OS=Tortoise microvirus 63 OX=2583169 PE=4 SV=1
MM1 pKa = 7.39YY2 pKa = 10.25SVCLDD7 pKa = 3.75DD8 pKa = 6.42GSIIGCGLGDD18 pKa = 4.33CYY20 pKa = 10.9DD21 pKa = 3.42VHH23 pKa = 8.53LLIPHH28 pKa = 7.04LMMDD32 pKa = 5.36AIDD35 pKa = 3.91NVLNGDD41 pKa = 3.9TMTLQDD47 pKa = 5.69FIRR50 pKa = 11.84LACAEE55 pKa = 4.1YY56 pKa = 11.13LFICGDD62 pKa = 3.5NYY64 pKa = 10.74VPKK67 pKa = 10.11TT68 pKa = 3.6
MM1 pKa = 7.39YY2 pKa = 10.25SVCLDD7 pKa = 3.75DD8 pKa = 6.42GSIIGCGLGDD18 pKa = 4.33CYY20 pKa = 10.9DD21 pKa = 3.42VHH23 pKa = 8.53LLIPHH28 pKa = 7.04LMMDD32 pKa = 5.36AIDD35 pKa = 3.91NVLNGDD41 pKa = 3.9TMTLQDD47 pKa = 5.69FIRR50 pKa = 11.84LACAEE55 pKa = 4.1YY56 pKa = 11.13LFICGDD62 pKa = 3.5NYY64 pKa = 10.74VPKK67 pKa = 10.11TT68 pKa = 3.6
Molecular weight: 7.52 kDa
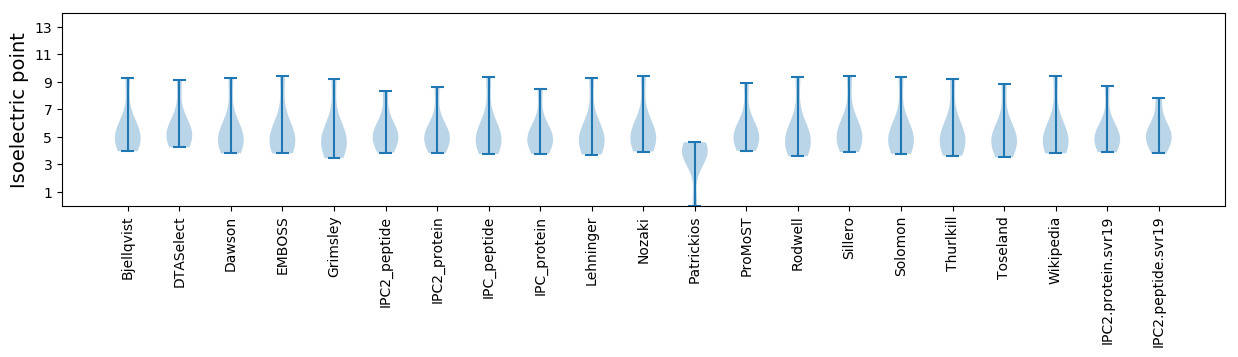
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7B6|A0A4P8W7B6_9VIRU Uncharacterized protein OS=Tortoise microvirus 63 OX=2583169 PE=4 SV=1
MM1 pKa = 7.4SCANPITITVKK12 pKa = 10.27GRR14 pKa = 11.84PVQVGCRR21 pKa = 11.84HH22 pKa = 6.08CLSCRR27 pKa = 11.84LDD29 pKa = 3.49YY30 pKa = 11.13QIALEE35 pKa = 4.07FAVKK39 pKa = 10.51HH40 pKa = 5.43EE41 pKa = 4.8LFDD44 pKa = 3.99IYY46 pKa = 11.03QSGLGACFATLTYY59 pKa = 10.51ADD61 pKa = 4.23SALPPHH67 pKa = 6.51GSLRR71 pKa = 11.84KK72 pKa = 9.48SDD74 pKa = 3.65FQNFLKK80 pKa = 10.59NVRR83 pKa = 11.84IQAKK87 pKa = 9.95RR88 pKa = 11.84KK89 pKa = 7.72GTLPPFKK96 pKa = 10.59YY97 pKa = 9.83LACGEE102 pKa = 4.31YY103 pKa = 10.34GDD105 pKa = 4.46KK106 pKa = 10.53FGRR109 pKa = 11.84PHH111 pKa = 5.25YY112 pKa = 10.13HH113 pKa = 6.39VIFLGVTDD121 pKa = 4.39ALFNQFIRR129 pKa = 11.84PLWKK133 pKa = 10.23HH134 pKa = 5.72GLCDD138 pKa = 4.08CSTLGSGAIRR148 pKa = 11.84YY149 pKa = 7.11VLKK152 pKa = 10.9YY153 pKa = 8.14CTKK156 pKa = 10.54SAMGSKK162 pKa = 10.03AKK164 pKa = 10.21EE165 pKa = 4.04MYY167 pKa = 10.28DD168 pKa = 3.65DD169 pKa = 4.01QSLTRR174 pKa = 11.84PFIVHH179 pKa = 6.05SKK181 pKa = 10.4RR182 pKa = 11.84LGFNWLIRR190 pKa = 11.84NRR192 pKa = 11.84DD193 pKa = 3.62DD194 pKa = 4.76LVANNFCYY202 pKa = 10.44LNKK205 pKa = 9.83GHH207 pKa = 7.02RR208 pKa = 11.84VPLPAYY214 pKa = 9.33YY215 pKa = 10.0RR216 pKa = 11.84RR217 pKa = 11.84QFDD220 pKa = 3.24IYY222 pKa = 10.89GTFCPLPQIQQLDD235 pKa = 3.89AEE237 pKa = 4.97AKK239 pKa = 7.73QHH241 pKa = 6.29GYY243 pKa = 9.49PSYY246 pKa = 10.82RR247 pKa = 11.84EE248 pKa = 3.74WSIQKK253 pKa = 10.22AFNRR257 pKa = 11.84EE258 pKa = 3.34QDD260 pKa = 3.13LTIAVRR266 pKa = 11.84NDD268 pKa = 4.52GIPVDD273 pKa = 4.8SLITPNRR280 pKa = 11.84GYY282 pKa = 10.99SPLDD286 pKa = 3.33TSIVPQVLNRR296 pKa = 11.84SS297 pKa = 3.62
MM1 pKa = 7.4SCANPITITVKK12 pKa = 10.27GRR14 pKa = 11.84PVQVGCRR21 pKa = 11.84HH22 pKa = 6.08CLSCRR27 pKa = 11.84LDD29 pKa = 3.49YY30 pKa = 11.13QIALEE35 pKa = 4.07FAVKK39 pKa = 10.51HH40 pKa = 5.43EE41 pKa = 4.8LFDD44 pKa = 3.99IYY46 pKa = 11.03QSGLGACFATLTYY59 pKa = 10.51ADD61 pKa = 4.23SALPPHH67 pKa = 6.51GSLRR71 pKa = 11.84KK72 pKa = 9.48SDD74 pKa = 3.65FQNFLKK80 pKa = 10.59NVRR83 pKa = 11.84IQAKK87 pKa = 9.95RR88 pKa = 11.84KK89 pKa = 7.72GTLPPFKK96 pKa = 10.59YY97 pKa = 9.83LACGEE102 pKa = 4.31YY103 pKa = 10.34GDD105 pKa = 4.46KK106 pKa = 10.53FGRR109 pKa = 11.84PHH111 pKa = 5.25YY112 pKa = 10.13HH113 pKa = 6.39VIFLGVTDD121 pKa = 4.39ALFNQFIRR129 pKa = 11.84PLWKK133 pKa = 10.23HH134 pKa = 5.72GLCDD138 pKa = 4.08CSTLGSGAIRR148 pKa = 11.84YY149 pKa = 7.11VLKK152 pKa = 10.9YY153 pKa = 8.14CTKK156 pKa = 10.54SAMGSKK162 pKa = 10.03AKK164 pKa = 10.21EE165 pKa = 4.04MYY167 pKa = 10.28DD168 pKa = 3.65DD169 pKa = 4.01QSLTRR174 pKa = 11.84PFIVHH179 pKa = 6.05SKK181 pKa = 10.4RR182 pKa = 11.84LGFNWLIRR190 pKa = 11.84NRR192 pKa = 11.84DD193 pKa = 3.62DD194 pKa = 4.76LVANNFCYY202 pKa = 10.44LNKK205 pKa = 9.83GHH207 pKa = 7.02RR208 pKa = 11.84VPLPAYY214 pKa = 9.33YY215 pKa = 10.0RR216 pKa = 11.84RR217 pKa = 11.84QFDD220 pKa = 3.24IYY222 pKa = 10.89GTFCPLPQIQQLDD235 pKa = 3.89AEE237 pKa = 4.97AKK239 pKa = 7.73QHH241 pKa = 6.29GYY243 pKa = 9.49PSYY246 pKa = 10.82RR247 pKa = 11.84EE248 pKa = 3.74WSIQKK253 pKa = 10.22AFNRR257 pKa = 11.84EE258 pKa = 3.34QDD260 pKa = 3.13LTIAVRR266 pKa = 11.84NDD268 pKa = 4.52GIPVDD273 pKa = 4.8SLITPNRR280 pKa = 11.84GYY282 pKa = 10.99SPLDD286 pKa = 3.33TSIVPQVLNRR296 pKa = 11.84SS297 pKa = 3.62
Molecular weight: 33.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1676 |
68 |
578 |
239.4 |
26.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.637 ± 1.396 | 1.372 ± 0.843 |
6.981 ± 0.811 | 4.057 ± 0.759 |
5.012 ± 0.574 | 6.146 ± 0.669 |
1.79 ± 0.294 | 4.893 ± 0.899 |
3.878 ± 1.222 | 8.652 ± 0.84 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.983 ± 0.458 | 4.654 ± 0.451 |
5.609 ± 0.709 | 5.43 ± 1.571 |
5.251 ± 0.468 | 9.069 ± 0.94 |
5.489 ± 0.923 | 5.489 ± 0.629 |
0.955 ± 0.143 | 4.654 ± 0.514 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
