
Hubei myriapoda virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
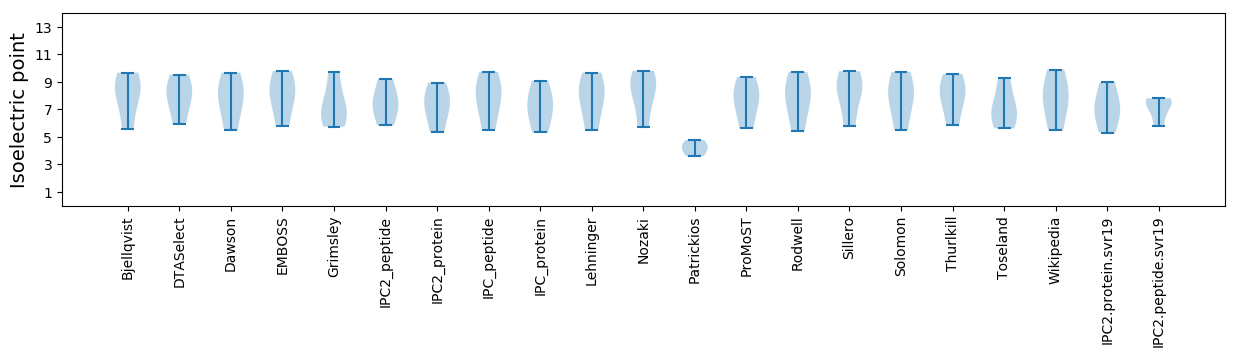
Average proteome isoelectric point is 7.1
Get precalculated fractions of proteins
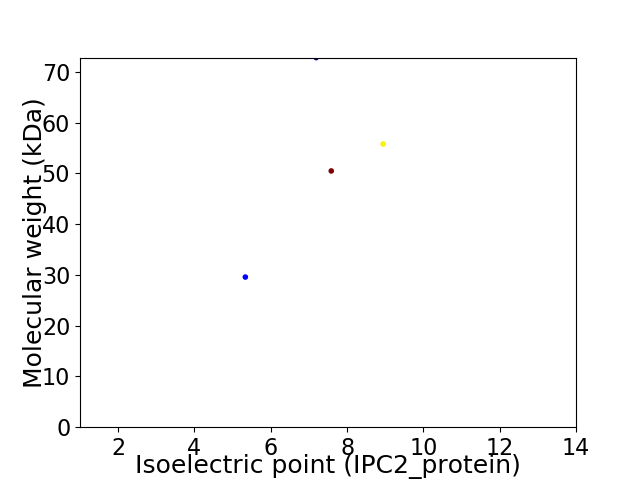
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEW9|A0A1L3KEW9_9VIRU Uncharacterized protein OS=Hubei myriapoda virus 9 OX=1922938 PE=4 SV=1
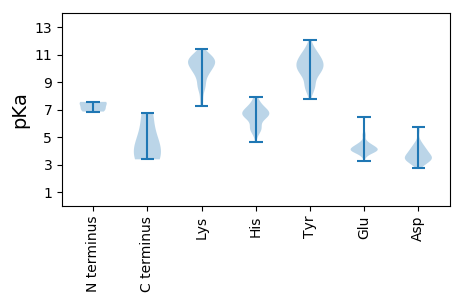
MM1 pKa = 7.52FYY3 pKa = 10.45IEE5 pKa = 5.03LLEE8 pKa = 4.33KK9 pKa = 10.88ALVTGSDD16 pKa = 3.24TGVVMGFNPFNGGYY30 pKa = 9.53KK31 pKa = 10.11HH32 pKa = 7.11FIRR35 pKa = 11.84SLPEE39 pKa = 3.61KK40 pKa = 10.57CLMLDD45 pKa = 3.18KK46 pKa = 11.16SSWDD50 pKa = 3.1WTVPTWLLSDD60 pKa = 4.62FYY62 pKa = 11.59QLLSNLGFGGCKK74 pKa = 9.49RR75 pKa = 11.84FHH77 pKa = 7.42RR78 pKa = 11.84WRR80 pKa = 11.84QLHH83 pKa = 6.43GIDD86 pKa = 4.55HH87 pKa = 6.53GFIPAMVEE95 pKa = 5.13GEE97 pKa = 4.39DD98 pKa = 3.78CDD100 pKa = 4.32LPAGIQVTGNFLTYY114 pKa = 10.13IGNSVMQLLLHH125 pKa = 6.76FMAEE129 pKa = 4.44DD130 pKa = 3.82LLHH133 pKa = 5.63EE134 pKa = 4.27QPGRR138 pKa = 11.84FYY140 pKa = 11.17CGGDD144 pKa = 3.5DD145 pKa = 5.37SIQSVPKK152 pKa = 10.64NYY154 pKa = 10.31DD155 pKa = 2.9GYY157 pKa = 11.64LEE159 pKa = 4.58FFQSVGLKK167 pKa = 9.84VRR169 pKa = 11.84GGISDD174 pKa = 2.86HH175 pKa = 6.69WEE177 pKa = 3.54FLGWKK182 pKa = 9.59FSKK185 pKa = 9.81EE186 pKa = 3.92RR187 pKa = 11.84KK188 pKa = 8.79PVPLYY193 pKa = 9.77SQKK196 pKa = 10.87HH197 pKa = 5.5LFNQQQLDD205 pKa = 3.84EE206 pKa = 5.62LIAPDD211 pKa = 4.73LLSSYY216 pKa = 10.62QLLYY220 pKa = 8.76TWEE223 pKa = 5.27PIMLNHH229 pKa = 6.92IKK231 pKa = 10.52SALSWIKK238 pKa = 10.61PEE240 pKa = 4.09FVRR243 pKa = 11.84SDD245 pKa = 2.97WYY247 pKa = 9.9LQCMWEE253 pKa = 4.19GLL255 pKa = 3.78
MM1 pKa = 7.52FYY3 pKa = 10.45IEE5 pKa = 5.03LLEE8 pKa = 4.33KK9 pKa = 10.88ALVTGSDD16 pKa = 3.24TGVVMGFNPFNGGYY30 pKa = 9.53KK31 pKa = 10.11HH32 pKa = 7.11FIRR35 pKa = 11.84SLPEE39 pKa = 3.61KK40 pKa = 10.57CLMLDD45 pKa = 3.18KK46 pKa = 11.16SSWDD50 pKa = 3.1WTVPTWLLSDD60 pKa = 4.62FYY62 pKa = 11.59QLLSNLGFGGCKK74 pKa = 9.49RR75 pKa = 11.84FHH77 pKa = 7.42RR78 pKa = 11.84WRR80 pKa = 11.84QLHH83 pKa = 6.43GIDD86 pKa = 4.55HH87 pKa = 6.53GFIPAMVEE95 pKa = 5.13GEE97 pKa = 4.39DD98 pKa = 3.78CDD100 pKa = 4.32LPAGIQVTGNFLTYY114 pKa = 10.13IGNSVMQLLLHH125 pKa = 6.76FMAEE129 pKa = 4.44DD130 pKa = 3.82LLHH133 pKa = 5.63EE134 pKa = 4.27QPGRR138 pKa = 11.84FYY140 pKa = 11.17CGGDD144 pKa = 3.5DD145 pKa = 5.37SIQSVPKK152 pKa = 10.64NYY154 pKa = 10.31DD155 pKa = 2.9GYY157 pKa = 11.64LEE159 pKa = 4.58FFQSVGLKK167 pKa = 9.84VRR169 pKa = 11.84GGISDD174 pKa = 2.86HH175 pKa = 6.69WEE177 pKa = 3.54FLGWKK182 pKa = 9.59FSKK185 pKa = 9.81EE186 pKa = 3.92RR187 pKa = 11.84KK188 pKa = 8.79PVPLYY193 pKa = 9.77SQKK196 pKa = 10.87HH197 pKa = 5.5LFNQQQLDD205 pKa = 3.84EE206 pKa = 5.62LIAPDD211 pKa = 4.73LLSSYY216 pKa = 10.62QLLYY220 pKa = 8.76TWEE223 pKa = 5.27PIMLNHH229 pKa = 6.92IKK231 pKa = 10.52SALSWIKK238 pKa = 10.61PEE240 pKa = 4.09FVRR243 pKa = 11.84SDD245 pKa = 2.97WYY247 pKa = 9.9LQCMWEE253 pKa = 4.19GLL255 pKa = 3.78
Molecular weight: 29.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEW5|A0A1L3KEW5_9VIRU RdRp OS=Hubei myriapoda virus 9 OX=1922938 PE=4 SV=1
MM1 pKa = 7.07SQSSNDD7 pKa = 3.09YY8 pKa = 11.0SMPKK12 pKa = 9.67VRR14 pKa = 11.84SLFDD18 pKa = 3.66LASDD22 pKa = 3.41QFLAVVALSWWLPFSLPWAIANLLLEE48 pKa = 5.2RR49 pKa = 11.84DD50 pKa = 3.86PGMPTAMLSYY60 pKa = 9.54PDD62 pKa = 4.24LVGTEE67 pKa = 4.49TKK69 pKa = 10.39RR70 pKa = 11.84KK71 pKa = 8.53PLRR74 pKa = 11.84LSTGVSRR81 pKa = 11.84SWRR84 pKa = 11.84HH85 pKa = 5.65CDD87 pKa = 2.78LCRR90 pKa = 11.84SIRR93 pKa = 11.84TNYY96 pKa = 9.43YY97 pKa = 8.66LPPRR101 pKa = 11.84FGVASCSEE109 pKa = 4.7CGCIWLSIPFHH120 pKa = 5.68GRR122 pKa = 11.84KK123 pKa = 9.31EE124 pKa = 3.88YY125 pKa = 10.53FLYY128 pKa = 10.77CKK130 pKa = 10.24VVQRR134 pKa = 11.84LLNRR138 pKa = 11.84GMQPLPLTRR147 pKa = 11.84DD148 pKa = 3.17ILRR151 pKa = 11.84RR152 pKa = 11.84PYY154 pKa = 10.53LVYY157 pKa = 9.08TGKK160 pKa = 9.76YY161 pKa = 8.87ARR163 pKa = 11.84FEE165 pKa = 4.28RR166 pKa = 11.84VVSTTILRR174 pKa = 11.84LAQMSDD180 pKa = 3.06SDD182 pKa = 3.66ICNTLWSLFSRR193 pKa = 11.84VRR195 pKa = 11.84HH196 pKa = 4.65TSAWGRR202 pKa = 11.84RR203 pKa = 11.84HH204 pKa = 6.79SGACDD209 pKa = 3.43RR210 pKa = 11.84KK211 pKa = 10.38CSVHH215 pKa = 6.66RR216 pKa = 11.84DD217 pKa = 3.24AVSFCNTGCSGCARR231 pKa = 11.84IQDD234 pKa = 3.48RR235 pKa = 11.84RR236 pKa = 11.84HH237 pKa = 5.28YY238 pKa = 10.35RR239 pKa = 11.84LSFLTNEE246 pKa = 4.54GFCLGTAAANDD257 pKa = 3.99CTAVRR262 pKa = 11.84CGTRR266 pKa = 11.84PEE268 pKa = 4.39HH269 pKa = 6.48FCTAVWRR276 pKa = 11.84SSGQAVDD283 pKa = 3.6ILWAIVASSKK293 pKa = 8.71VVRR296 pKa = 11.84IPEE299 pKa = 3.98ILWIHH304 pKa = 5.47TKK306 pKa = 10.66GGIWGTNCSWQTSCSYY322 pKa = 11.17ARR324 pKa = 11.84WLRR327 pKa = 11.84GQIQHH332 pKa = 7.17LDD334 pKa = 3.3RR335 pKa = 11.84DD336 pKa = 4.28GLHH339 pKa = 7.05LILFRR344 pKa = 11.84PAEE347 pKa = 4.11QIGLAILPEE356 pKa = 4.53DD357 pKa = 4.19PNPVEE362 pKa = 4.04VQPGIICEE370 pKa = 3.97YY371 pKa = 9.99GRR373 pKa = 11.84VSRR376 pKa = 11.84EE377 pKa = 3.66LGSKK381 pKa = 10.17RR382 pKa = 11.84KK383 pKa = 7.56EE384 pKa = 4.23TQGLARR390 pKa = 11.84YY391 pKa = 8.28GRR393 pKa = 11.84QRR395 pKa = 11.84LLPRR399 pKa = 11.84TWWQDD404 pKa = 3.4PIDD407 pKa = 4.11RR408 pKa = 11.84PIYY411 pKa = 10.83VNGCYY416 pKa = 9.76ARR418 pKa = 11.84CIGLVPHH425 pKa = 6.55RR426 pKa = 11.84TFGPSFWRR434 pKa = 11.84LGRR437 pKa = 11.84LHH439 pKa = 7.09GPLFSGVRR447 pKa = 11.84RR448 pKa = 11.84EE449 pKa = 3.99SEE451 pKa = 3.77MRR453 pKa = 11.84RR454 pKa = 11.84LRR456 pKa = 11.84DD457 pKa = 3.52RR458 pKa = 11.84IQRR461 pKa = 11.84DD462 pKa = 3.39RR463 pKa = 11.84EE464 pKa = 4.06RR465 pKa = 11.84EE466 pKa = 3.9RR467 pKa = 11.84SSLPSSAEE475 pKa = 3.89AQHH478 pKa = 6.45SEE480 pKa = 3.95NDD482 pKa = 3.57SNQIGG487 pKa = 3.38
MM1 pKa = 7.07SQSSNDD7 pKa = 3.09YY8 pKa = 11.0SMPKK12 pKa = 9.67VRR14 pKa = 11.84SLFDD18 pKa = 3.66LASDD22 pKa = 3.41QFLAVVALSWWLPFSLPWAIANLLLEE48 pKa = 5.2RR49 pKa = 11.84DD50 pKa = 3.86PGMPTAMLSYY60 pKa = 9.54PDD62 pKa = 4.24LVGTEE67 pKa = 4.49TKK69 pKa = 10.39RR70 pKa = 11.84KK71 pKa = 8.53PLRR74 pKa = 11.84LSTGVSRR81 pKa = 11.84SWRR84 pKa = 11.84HH85 pKa = 5.65CDD87 pKa = 2.78LCRR90 pKa = 11.84SIRR93 pKa = 11.84TNYY96 pKa = 9.43YY97 pKa = 8.66LPPRR101 pKa = 11.84FGVASCSEE109 pKa = 4.7CGCIWLSIPFHH120 pKa = 5.68GRR122 pKa = 11.84KK123 pKa = 9.31EE124 pKa = 3.88YY125 pKa = 10.53FLYY128 pKa = 10.77CKK130 pKa = 10.24VVQRR134 pKa = 11.84LLNRR138 pKa = 11.84GMQPLPLTRR147 pKa = 11.84DD148 pKa = 3.17ILRR151 pKa = 11.84RR152 pKa = 11.84PYY154 pKa = 10.53LVYY157 pKa = 9.08TGKK160 pKa = 9.76YY161 pKa = 8.87ARR163 pKa = 11.84FEE165 pKa = 4.28RR166 pKa = 11.84VVSTTILRR174 pKa = 11.84LAQMSDD180 pKa = 3.06SDD182 pKa = 3.66ICNTLWSLFSRR193 pKa = 11.84VRR195 pKa = 11.84HH196 pKa = 4.65TSAWGRR202 pKa = 11.84RR203 pKa = 11.84HH204 pKa = 6.79SGACDD209 pKa = 3.43RR210 pKa = 11.84KK211 pKa = 10.38CSVHH215 pKa = 6.66RR216 pKa = 11.84DD217 pKa = 3.24AVSFCNTGCSGCARR231 pKa = 11.84IQDD234 pKa = 3.48RR235 pKa = 11.84RR236 pKa = 11.84HH237 pKa = 5.28YY238 pKa = 10.35RR239 pKa = 11.84LSFLTNEE246 pKa = 4.54GFCLGTAAANDD257 pKa = 3.99CTAVRR262 pKa = 11.84CGTRR266 pKa = 11.84PEE268 pKa = 4.39HH269 pKa = 6.48FCTAVWRR276 pKa = 11.84SSGQAVDD283 pKa = 3.6ILWAIVASSKK293 pKa = 8.71VVRR296 pKa = 11.84IPEE299 pKa = 3.98ILWIHH304 pKa = 5.47TKK306 pKa = 10.66GGIWGTNCSWQTSCSYY322 pKa = 11.17ARR324 pKa = 11.84WLRR327 pKa = 11.84GQIQHH332 pKa = 7.17LDD334 pKa = 3.3RR335 pKa = 11.84DD336 pKa = 4.28GLHH339 pKa = 7.05LILFRR344 pKa = 11.84PAEE347 pKa = 4.11QIGLAILPEE356 pKa = 4.53DD357 pKa = 4.19PNPVEE362 pKa = 4.04VQPGIICEE370 pKa = 3.97YY371 pKa = 9.99GRR373 pKa = 11.84VSRR376 pKa = 11.84EE377 pKa = 3.66LGSKK381 pKa = 10.17RR382 pKa = 11.84KK383 pKa = 7.56EE384 pKa = 4.23TQGLARR390 pKa = 11.84YY391 pKa = 8.28GRR393 pKa = 11.84QRR395 pKa = 11.84LLPRR399 pKa = 11.84TWWQDD404 pKa = 3.4PIDD407 pKa = 4.11RR408 pKa = 11.84PIYY411 pKa = 10.83VNGCYY416 pKa = 9.76ARR418 pKa = 11.84CIGLVPHH425 pKa = 6.55RR426 pKa = 11.84TFGPSFWRR434 pKa = 11.84LGRR437 pKa = 11.84LHH439 pKa = 7.09GPLFSGVRR447 pKa = 11.84RR448 pKa = 11.84EE449 pKa = 3.99SEE451 pKa = 3.77MRR453 pKa = 11.84RR454 pKa = 11.84LRR456 pKa = 11.84DD457 pKa = 3.52RR458 pKa = 11.84IQRR461 pKa = 11.84DD462 pKa = 3.39RR463 pKa = 11.84EE464 pKa = 4.06RR465 pKa = 11.84EE466 pKa = 3.9RR467 pKa = 11.84SSLPSSAEE475 pKa = 3.89AQHH478 pKa = 6.45SEE480 pKa = 3.95NDD482 pKa = 3.57SNQIGG487 pKa = 3.38
Molecular weight: 55.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1870 |
255 |
667 |
467.5 |
52.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.471 ± 0.765 | 2.781 ± 0.513 |
4.973 ± 0.291 | 4.492 ± 0.379 |
3.797 ± 0.51 | 7.754 ± 0.285 |
2.246 ± 0.281 | 4.652 ± 0.187 |
4.439 ± 0.776 | 8.93 ± 0.854 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.032 ± 0.251 | 2.674 ± 0.473 |
6.364 ± 0.421 | 4.545 ± 0.419 |
6.471 ± 1.753 | 9.626 ± 1.089 |
5.294 ± 0.63 | 6.417 ± 0.498 |
2.674 ± 0.313 | 3.369 ± 0.221 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
