
Tomato pseudo-curly top virus (TPCTV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Topocuvirus
Average proteome isoelectric point is 7.3
Get precalculated fractions of proteins
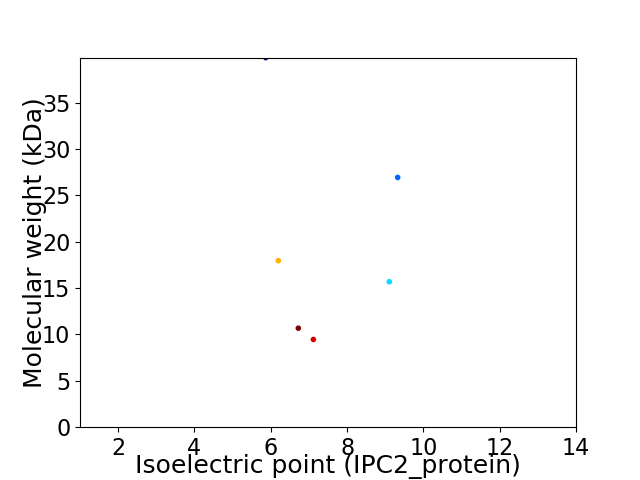
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q88889|C2_TPCTV Protein C2 OS=Tomato pseudo-curly top virus OX=49267 GN=C2 PE=3 SV=1
MM1 pKa = 7.85PSNPKK6 pKa = 9.99RR7 pKa = 11.84FQIAAKK13 pKa = 10.09NYY15 pKa = 8.13FLTYY19 pKa = 9.42PNCSLSKK26 pKa = 11.06EE27 pKa = 4.09EE28 pKa = 6.13ALDD31 pKa = 3.41QLQRR35 pKa = 11.84LQTPTNKK42 pKa = 10.17KK43 pKa = 7.7YY44 pKa = 10.69IKK46 pKa = 9.88VARR49 pKa = 11.84EE50 pKa = 3.51LHH52 pKa = 6.43EE53 pKa = 4.62NGEE56 pKa = 4.28PHH58 pKa = 6.66LHH60 pKa = 6.03VLIQFEE66 pKa = 5.08GKK68 pKa = 10.06FNCKK72 pKa = 7.32NQRR75 pKa = 11.84FFDD78 pKa = 4.05LVSPTRR84 pKa = 11.84STHH87 pKa = 4.78FHH89 pKa = 6.87PNIQGAKK96 pKa = 9.58SSSDD100 pKa = 3.24VNSYY104 pKa = 9.77VDD106 pKa = 3.82KK107 pKa = 11.49DD108 pKa = 3.28GDD110 pKa = 3.95TIEE113 pKa = 4.07WGEE116 pKa = 3.82FQIDD120 pKa = 3.13ARR122 pKa = 11.84SARR125 pKa = 11.84GGQQTANDD133 pKa = 3.74EE134 pKa = 4.18CAEE137 pKa = 3.93ALNRR141 pKa = 11.84SSKK144 pKa = 10.97EE145 pKa = 3.45EE146 pKa = 3.49ALQIIKK152 pKa = 10.52EE153 pKa = 4.07KK154 pKa = 10.65LPKK157 pKa = 10.5DD158 pKa = 3.74FLFCYY163 pKa = 10.31HH164 pKa = 6.57NLVSNLDD171 pKa = 4.14RR172 pKa = 11.84IFTPAPTPFVPPFQLSSFTNVPEE195 pKa = 5.65DD196 pKa = 3.6MQEE199 pKa = 3.7WADD202 pKa = 3.87DD203 pKa = 3.96YY204 pKa = 11.59FGVSAAARR212 pKa = 11.84PMRR215 pKa = 11.84YY216 pKa = 9.6KK217 pKa = 10.74SIIIEE222 pKa = 4.3GEE224 pKa = 3.96SRR226 pKa = 11.84TGKK229 pKa = 8.58TMWARR234 pKa = 11.84SLGPHH239 pKa = 6.57NYY241 pKa = 10.17LSGHH245 pKa = 6.9LDD247 pKa = 3.48FNSRR251 pKa = 11.84VYY253 pKa = 11.25SNSALYY259 pKa = 10.53NVIDD263 pKa = 4.88DD264 pKa = 4.15VTPHH268 pKa = 5.44YY269 pKa = 10.89LKK271 pKa = 10.69LKK273 pKa = 9.09HH274 pKa = 5.7WKK276 pKa = 9.57EE277 pKa = 4.04LIGAQRR283 pKa = 11.84DD284 pKa = 3.53WQSNCKK290 pKa = 9.41YY291 pKa = 10.21GKK293 pKa = 9.18PVQIKK298 pKa = 10.19GGIPSIVLCNPGGDD312 pKa = 3.38TSFQDD317 pKa = 4.37FLDD320 pKa = 4.08KK321 pKa = 11.3EE322 pKa = 4.26EE323 pKa = 4.61NEE325 pKa = 4.27ALKK328 pKa = 10.83DD329 pKa = 3.27WTLYY333 pKa = 10.13NAVFIKK339 pKa = 9.85LTEE342 pKa = 3.99PLYY345 pKa = 11.14DD346 pKa = 3.49GTVV349 pKa = 2.82
MM1 pKa = 7.85PSNPKK6 pKa = 9.99RR7 pKa = 11.84FQIAAKK13 pKa = 10.09NYY15 pKa = 8.13FLTYY19 pKa = 9.42PNCSLSKK26 pKa = 11.06EE27 pKa = 4.09EE28 pKa = 6.13ALDD31 pKa = 3.41QLQRR35 pKa = 11.84LQTPTNKK42 pKa = 10.17KK43 pKa = 7.7YY44 pKa = 10.69IKK46 pKa = 9.88VARR49 pKa = 11.84EE50 pKa = 3.51LHH52 pKa = 6.43EE53 pKa = 4.62NGEE56 pKa = 4.28PHH58 pKa = 6.66LHH60 pKa = 6.03VLIQFEE66 pKa = 5.08GKK68 pKa = 10.06FNCKK72 pKa = 7.32NQRR75 pKa = 11.84FFDD78 pKa = 4.05LVSPTRR84 pKa = 11.84STHH87 pKa = 4.78FHH89 pKa = 6.87PNIQGAKK96 pKa = 9.58SSSDD100 pKa = 3.24VNSYY104 pKa = 9.77VDD106 pKa = 3.82KK107 pKa = 11.49DD108 pKa = 3.28GDD110 pKa = 3.95TIEE113 pKa = 4.07WGEE116 pKa = 3.82FQIDD120 pKa = 3.13ARR122 pKa = 11.84SARR125 pKa = 11.84GGQQTANDD133 pKa = 3.74EE134 pKa = 4.18CAEE137 pKa = 3.93ALNRR141 pKa = 11.84SSKK144 pKa = 10.97EE145 pKa = 3.45EE146 pKa = 3.49ALQIIKK152 pKa = 10.52EE153 pKa = 4.07KK154 pKa = 10.65LPKK157 pKa = 10.5DD158 pKa = 3.74FLFCYY163 pKa = 10.31HH164 pKa = 6.57NLVSNLDD171 pKa = 4.14RR172 pKa = 11.84IFTPAPTPFVPPFQLSSFTNVPEE195 pKa = 5.65DD196 pKa = 3.6MQEE199 pKa = 3.7WADD202 pKa = 3.87DD203 pKa = 3.96YY204 pKa = 11.59FGVSAAARR212 pKa = 11.84PMRR215 pKa = 11.84YY216 pKa = 9.6KK217 pKa = 10.74SIIIEE222 pKa = 4.3GEE224 pKa = 3.96SRR226 pKa = 11.84TGKK229 pKa = 8.58TMWARR234 pKa = 11.84SLGPHH239 pKa = 6.57NYY241 pKa = 10.17LSGHH245 pKa = 6.9LDD247 pKa = 3.48FNSRR251 pKa = 11.84VYY253 pKa = 11.25SNSALYY259 pKa = 10.53NVIDD263 pKa = 4.88DD264 pKa = 4.15VTPHH268 pKa = 5.44YY269 pKa = 10.89LKK271 pKa = 10.69LKK273 pKa = 9.09HH274 pKa = 5.7WKK276 pKa = 9.57EE277 pKa = 4.04LIGAQRR283 pKa = 11.84DD284 pKa = 3.53WQSNCKK290 pKa = 9.41YY291 pKa = 10.21GKK293 pKa = 9.18PVQIKK298 pKa = 10.19GGIPSIVLCNPGGDD312 pKa = 3.38TSFQDD317 pKa = 4.37FLDD320 pKa = 4.08KK321 pKa = 11.3EE322 pKa = 4.26EE323 pKa = 4.61NEE325 pKa = 4.27ALKK328 pKa = 10.83DD329 pKa = 3.27WTLYY333 pKa = 10.13NAVFIKK339 pKa = 9.85LTEE342 pKa = 3.99PLYY345 pKa = 11.14DD346 pKa = 3.49GTVV349 pKa = 2.82
Molecular weight: 39.84 kDa
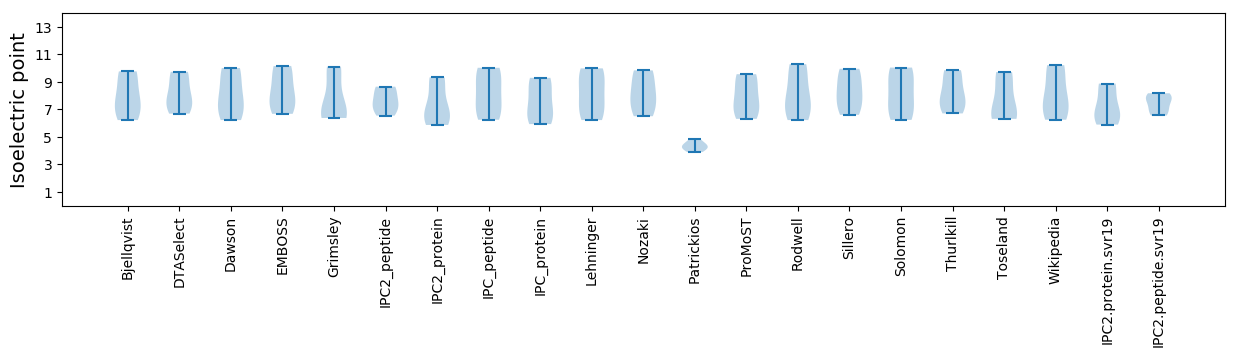
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q88891|AC4_TPCTV Protein C4 OS=Tomato pseudo-curly top virus OX=49267 GN=C4 PE=3 SV=1
MM1 pKa = 8.27DD2 pKa = 4.3SRR4 pKa = 11.84TGEE7 pKa = 4.27LLTYY11 pKa = 7.18TQCQTGCYY19 pKa = 9.59FDD21 pKa = 6.0DD22 pKa = 4.24IKK24 pKa = 11.58NPIYY28 pKa = 10.53FRR30 pKa = 11.84LQNTTTMATGNKK42 pKa = 9.5IITLQIRR49 pKa = 11.84ANHH52 pKa = 6.09NLRR55 pKa = 11.84KK56 pKa = 10.04VLGLQICWLNLVVLSVSPRR75 pKa = 11.84LSGEE79 pKa = 3.86ALLDD83 pKa = 3.32RR84 pKa = 11.84FKK86 pKa = 11.18FHH88 pKa = 6.98VMKK91 pKa = 10.55HH92 pKa = 5.2LNNLGVISINNVIRR106 pKa = 11.84SINHH110 pKa = 6.77FIDD113 pKa = 5.26LFDD116 pKa = 3.38QRR118 pKa = 11.84VFYY121 pKa = 10.9APSFYY126 pKa = 11.26YY127 pKa = 9.76NIKK130 pKa = 8.76MRR132 pKa = 11.84LYY134 pKa = 10.62
MM1 pKa = 8.27DD2 pKa = 4.3SRR4 pKa = 11.84TGEE7 pKa = 4.27LLTYY11 pKa = 7.18TQCQTGCYY19 pKa = 9.59FDD21 pKa = 6.0DD22 pKa = 4.24IKK24 pKa = 11.58NPIYY28 pKa = 10.53FRR30 pKa = 11.84LQNTTTMATGNKK42 pKa = 9.5IITLQIRR49 pKa = 11.84ANHH52 pKa = 6.09NLRR55 pKa = 11.84KK56 pKa = 10.04VLGLQICWLNLVVLSVSPRR75 pKa = 11.84LSGEE79 pKa = 3.86ALLDD83 pKa = 3.32RR84 pKa = 11.84FKK86 pKa = 11.18FHH88 pKa = 6.98VMKK91 pKa = 10.55HH92 pKa = 5.2LNNLGVISINNVIRR106 pKa = 11.84SINHH110 pKa = 6.77FIDD113 pKa = 5.26LFDD116 pKa = 3.38QRR118 pKa = 11.84VFYY121 pKa = 10.9APSFYY126 pKa = 11.26YY127 pKa = 9.76NIKK130 pKa = 8.76MRR132 pKa = 11.84LYY134 pKa = 10.62
Molecular weight: 15.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1065 |
85 |
349 |
177.5 |
20.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.822 ± 0.754 | 2.723 ± 0.563 |
4.789 ± 0.553 | 5.164 ± 1.327 |
4.038 ± 0.742 | 6.009 ± 0.541 |
2.347 ± 0.526 | 6.009 ± 0.582 |
6.197 ± 0.671 | 8.451 ± 0.845 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.784 ± 0.46 | 6.103 ± 0.738 |
5.258 ± 0.856 | 3.944 ± 0.409 |
6.009 ± 0.602 | 8.545 ± 0.906 |
6.291 ± 0.943 | 4.789 ± 1.026 |
1.315 ± 0.213 | 4.413 ± 0.675 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
