
Rhodosporidium toruloides (Yeast) (Rhodotorula gracilis)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota;
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
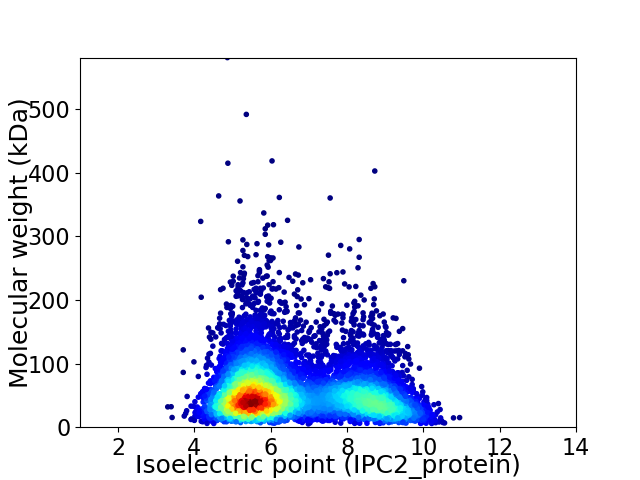
Virtual 2D-PAGE plot for 7496 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K3C6Z5|A0A0K3C6Z5_RHOTO FGENESH: predicted gene_1.91 protein (Fragment) OS=Rhodosporidium toruloides OX=5286 GN=FGENESH: predicted gene_1.91 PE=4 SV=1
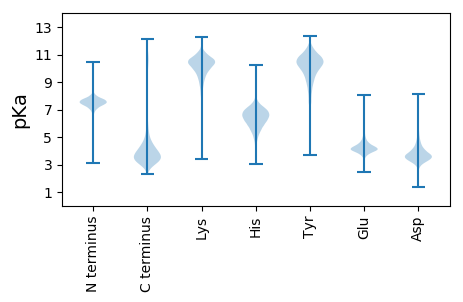
MM1 pKa = 7.62GLLDD5 pKa = 3.84KK6 pKa = 11.3AKK8 pKa = 10.59DD9 pKa = 3.99AISHH13 pKa = 6.64HH14 pKa = 5.59GQNKK18 pKa = 9.8NSGSDD23 pKa = 3.27NYY25 pKa = 11.29NDD27 pKa = 3.38NTTGDD32 pKa = 4.1SYY34 pKa = 12.09NSGNQSYY41 pKa = 8.93GQQDD45 pKa = 3.47SSFGGASSGFDD56 pKa = 3.33STSDD60 pKa = 3.25RR61 pKa = 11.84YY62 pKa = 10.82GSSDD66 pKa = 3.38NQGSGGFTSGDD77 pKa = 3.42NNNNQDD83 pKa = 3.78DD84 pKa = 4.25SFSNSRR90 pKa = 11.84RR91 pKa = 11.84AGAAEE96 pKa = 3.9GDD98 pKa = 4.23SYY100 pKa = 11.98GGASGGFDD108 pKa = 3.33STSDD112 pKa = 3.4RR113 pKa = 11.84YY114 pKa = 11.02GSGGNDD120 pKa = 3.36DD121 pKa = 4.72SYY123 pKa = 12.08GSGGAGRR130 pKa = 11.84SSGLGANDD138 pKa = 3.44MTSGRR143 pKa = 11.84QSDD146 pKa = 4.08FDD148 pKa = 4.44SGSQSYY154 pKa = 10.94GDD156 pKa = 3.56NSYY159 pKa = 11.36GSNRR163 pKa = 11.84ASGFDD168 pKa = 3.57DD169 pKa = 4.4ASNISDD175 pKa = 3.28TGNRR179 pKa = 11.84LDD181 pKa = 3.38SSYY184 pKa = 11.58GGGSGRR190 pKa = 11.84GTDD193 pKa = 4.27FQASGGGSAYY203 pKa = 10.51GDD205 pKa = 3.64GQTDD209 pKa = 3.82FGGAGRR215 pKa = 11.84NNDD218 pKa = 3.2GQYY221 pKa = 11.09GDD223 pKa = 4.04SSFGGSSGNQGYY235 pKa = 8.21GQDD238 pKa = 3.32QTQYY242 pKa = 11.18SGGGNQYY249 pKa = 10.63GGEE252 pKa = 3.97RR253 pKa = 11.84DD254 pKa = 3.52NYY256 pKa = 9.33PP257 pKa = 3.18
MM1 pKa = 7.62GLLDD5 pKa = 3.84KK6 pKa = 11.3AKK8 pKa = 10.59DD9 pKa = 3.99AISHH13 pKa = 6.64HH14 pKa = 5.59GQNKK18 pKa = 9.8NSGSDD23 pKa = 3.27NYY25 pKa = 11.29NDD27 pKa = 3.38NTTGDD32 pKa = 4.1SYY34 pKa = 12.09NSGNQSYY41 pKa = 8.93GQQDD45 pKa = 3.47SSFGGASSGFDD56 pKa = 3.33STSDD60 pKa = 3.25RR61 pKa = 11.84YY62 pKa = 10.82GSSDD66 pKa = 3.38NQGSGGFTSGDD77 pKa = 3.42NNNNQDD83 pKa = 3.78DD84 pKa = 4.25SFSNSRR90 pKa = 11.84RR91 pKa = 11.84AGAAEE96 pKa = 3.9GDD98 pKa = 4.23SYY100 pKa = 11.98GGASGGFDD108 pKa = 3.33STSDD112 pKa = 3.4RR113 pKa = 11.84YY114 pKa = 11.02GSGGNDD120 pKa = 3.36DD121 pKa = 4.72SYY123 pKa = 12.08GSGGAGRR130 pKa = 11.84SSGLGANDD138 pKa = 3.44MTSGRR143 pKa = 11.84QSDD146 pKa = 4.08FDD148 pKa = 4.44SGSQSYY154 pKa = 10.94GDD156 pKa = 3.56NSYY159 pKa = 11.36GSNRR163 pKa = 11.84ASGFDD168 pKa = 3.57DD169 pKa = 4.4ASNISDD175 pKa = 3.28TGNRR179 pKa = 11.84LDD181 pKa = 3.38SSYY184 pKa = 11.58GGGSGRR190 pKa = 11.84GTDD193 pKa = 4.27FQASGGGSAYY203 pKa = 10.51GDD205 pKa = 3.64GQTDD209 pKa = 3.82FGGAGRR215 pKa = 11.84NNDD218 pKa = 3.2GQYY221 pKa = 11.09GDD223 pKa = 4.04SSFGGSSGNQGYY235 pKa = 8.21GQDD238 pKa = 3.32QTQYY242 pKa = 11.18SGGGNQYY249 pKa = 10.63GGEE252 pKa = 3.97RR253 pKa = 11.84DD254 pKa = 3.52NYY256 pKa = 9.33PP257 pKa = 3.18
Molecular weight: 25.85 kDa
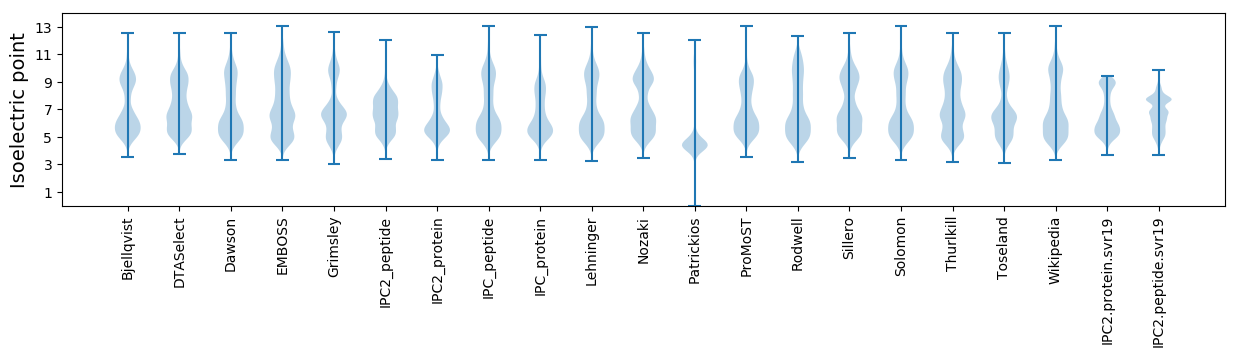
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K3C6B7|A0A0K3C6B7_RHOTO BY PROTMAP: gi|342319937|gb|EGU11882.1| Proteophosphoglycan 5 [Rhodotorula glutinis ATCC 204091] OS=Rhodosporidium toruloides OX=5286 GN=FGENESH: predicted gene_2.254 PE=4 SV=1
MM1 pKa = 7.36MLNALRR7 pKa = 11.84SASRR11 pKa = 11.84TLSRR15 pKa = 11.84TVQIAGARR23 pKa = 11.84SPLAAPSHH31 pKa = 6.23SRR33 pKa = 11.84LISTSSSALSPILSTAASRR52 pKa = 11.84LTSAAARR59 pKa = 11.84PQTSLLQRR67 pKa = 11.84EE68 pKa = 4.43LAQAGTRR75 pKa = 11.84PGFWHH80 pKa = 6.96TSPSPLSSAVQQQVRR95 pKa = 11.84GYY97 pKa = 11.12KK98 pKa = 8.93MPKK101 pKa = 8.81CMRR104 pKa = 11.84RR105 pKa = 11.84KK106 pKa = 9.95ASPLARR112 pKa = 11.84NGGKK116 pKa = 10.51GKK118 pKa = 6.7TARR121 pKa = 11.84RR122 pKa = 11.84KK123 pKa = 10.55GMLKK127 pKa = 10.08AKK129 pKa = 9.45RR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84MRR134 pKa = 11.84IKK136 pKa = 10.51KK137 pKa = 10.05INN139 pKa = 3.2
MM1 pKa = 7.36MLNALRR7 pKa = 11.84SASRR11 pKa = 11.84TLSRR15 pKa = 11.84TVQIAGARR23 pKa = 11.84SPLAAPSHH31 pKa = 6.23SRR33 pKa = 11.84LISTSSSALSPILSTAASRR52 pKa = 11.84LTSAAARR59 pKa = 11.84PQTSLLQRR67 pKa = 11.84EE68 pKa = 4.43LAQAGTRR75 pKa = 11.84PGFWHH80 pKa = 6.96TSPSPLSSAVQQQVRR95 pKa = 11.84GYY97 pKa = 11.12KK98 pKa = 8.93MPKK101 pKa = 8.81CMRR104 pKa = 11.84RR105 pKa = 11.84KK106 pKa = 9.95ASPLARR112 pKa = 11.84NGGKK116 pKa = 10.51GKK118 pKa = 6.7TARR121 pKa = 11.84RR122 pKa = 11.84KK123 pKa = 10.55GMLKK127 pKa = 10.08AKK129 pKa = 9.45RR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84MRR134 pKa = 11.84IKK136 pKa = 10.51KK137 pKa = 10.05INN139 pKa = 3.2
Molecular weight: 15.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4252312 |
50 |
5355 |
567.3 |
61.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.691 ± 0.031 | 0.998 ± 0.012 |
5.499 ± 0.019 | 6.381 ± 0.027 |
3.41 ± 0.016 | 7.116 ± 0.027 |
2.207 ± 0.01 | 3.596 ± 0.02 |
4.417 ± 0.023 | 9.372 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.668 ± 0.008 | 2.559 ± 0.015 |
7.158 ± 0.038 | 3.63 ± 0.02 |
6.738 ± 0.025 | 9.071 ± 0.039 |
5.735 ± 0.019 | 6.19 ± 0.019 |
1.287 ± 0.009 | 2.276 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
