
Streptococcus phage CHPC925
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Moineauvirus; unclassified Moineauvirus
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
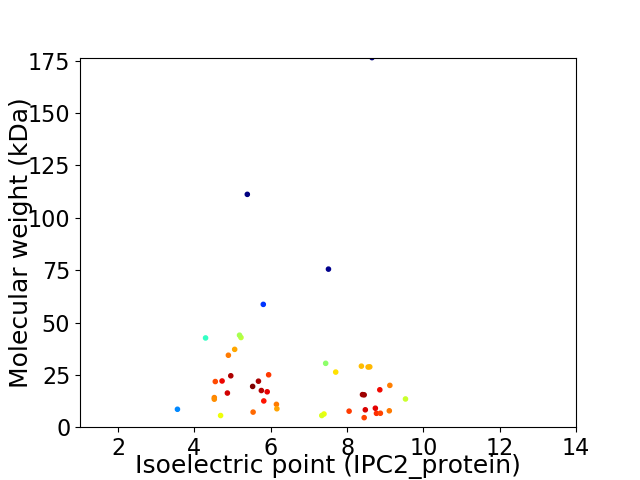
Virtual 2D-PAGE plot for 45 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
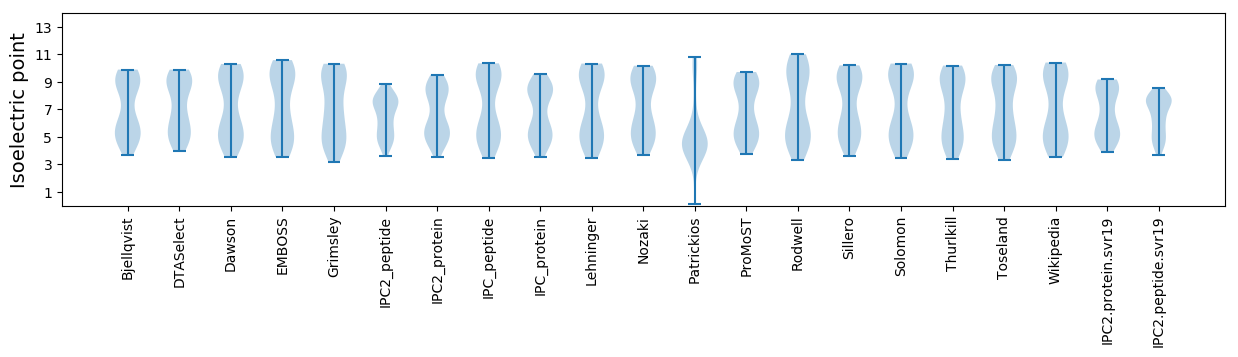
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G8F7R1|A0A3G8F7R1_9CAUD Uncharacterized protein OS=Streptococcus phage CHPC925 OX=2365047 GN=CHPC925_0026 PE=4 SV=1
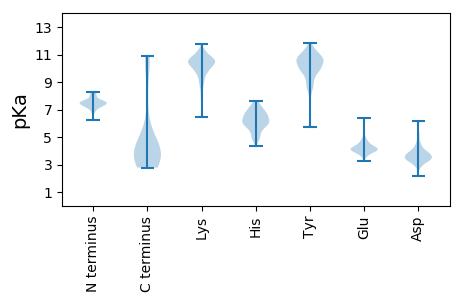
MM1 pKa = 7.11VNWVDD6 pKa = 4.52ADD8 pKa = 4.28GNDD11 pKa = 4.07LPDD14 pKa = 5.09GADD17 pKa = 3.18QDD19 pKa = 4.63FKK21 pKa = 11.75AGMFFSFAGDD31 pKa = 3.45EE32 pKa = 4.32VNITDD37 pKa = 4.04TGTGGYY43 pKa = 9.64YY44 pKa = 10.32GGYY47 pKa = 7.71YY48 pKa = 8.88WRR50 pKa = 11.84LFEE53 pKa = 4.82FGQFGTVWLSCWNKK67 pKa = 10.66DD68 pKa = 3.89DD69 pKa = 5.14LVNYY73 pKa = 7.38YY74 pKa = 8.26QQ75 pKa = 4.22
MM1 pKa = 7.11VNWVDD6 pKa = 4.52ADD8 pKa = 4.28GNDD11 pKa = 4.07LPDD14 pKa = 5.09GADD17 pKa = 3.18QDD19 pKa = 4.63FKK21 pKa = 11.75AGMFFSFAGDD31 pKa = 3.45EE32 pKa = 4.32VNITDD37 pKa = 4.04TGTGGYY43 pKa = 9.64YY44 pKa = 10.32GGYY47 pKa = 7.71YY48 pKa = 8.88WRR50 pKa = 11.84LFEE53 pKa = 4.82FGQFGTVWLSCWNKK67 pKa = 10.66DD68 pKa = 3.89DD69 pKa = 5.14LVNYY73 pKa = 7.38YY74 pKa = 8.26QQ75 pKa = 4.22
Molecular weight: 8.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G8F7X5|A0A3G8F7X5_9CAUD Uncharacterized protein OS=Streptococcus phage CHPC925 OX=2365047 GN=CHPC925_0018 PE=4 SV=1
MM1 pKa = 7.36ARR3 pKa = 11.84VRR5 pKa = 11.84YY6 pKa = 9.32LPSDD10 pKa = 3.79FRR12 pKa = 11.84YY13 pKa = 9.99KK14 pKa = 10.95ADD16 pKa = 3.48FGTYY20 pKa = 9.38QSTPNKK26 pKa = 8.16FTGVSVPKK34 pKa = 10.14FVKK37 pKa = 10.38QFTLHH42 pKa = 5.9YY43 pKa = 9.16KK44 pKa = 9.46PHH46 pKa = 6.1TRR48 pKa = 11.84TLNQEE53 pKa = 3.83YY54 pKa = 10.01LAQQNGEE61 pKa = 3.75SDD63 pKa = 3.27TRR65 pKa = 11.84VIVIRR70 pKa = 11.84HH71 pKa = 4.59NVKK74 pKa = 10.47VIEE77 pKa = 4.53GQAVVLNGTQYY88 pKa = 11.37DD89 pKa = 3.79IVRR92 pKa = 11.84VSSNEE97 pKa = 3.55NFGLNRR103 pKa = 11.84YY104 pKa = 9.75DD105 pKa = 3.94YY106 pKa = 10.41LTLKK110 pKa = 9.73KK111 pKa = 9.68HH112 pKa = 5.98KK113 pKa = 10.48KK114 pKa = 9.32IGG116 pKa = 3.46
MM1 pKa = 7.36ARR3 pKa = 11.84VRR5 pKa = 11.84YY6 pKa = 9.32LPSDD10 pKa = 3.79FRR12 pKa = 11.84YY13 pKa = 9.99KK14 pKa = 10.95ADD16 pKa = 3.48FGTYY20 pKa = 9.38QSTPNKK26 pKa = 8.16FTGVSVPKK34 pKa = 10.14FVKK37 pKa = 10.38QFTLHH42 pKa = 5.9YY43 pKa = 9.16KK44 pKa = 9.46PHH46 pKa = 6.1TRR48 pKa = 11.84TLNQEE53 pKa = 3.83YY54 pKa = 10.01LAQQNGEE61 pKa = 3.75SDD63 pKa = 3.27TRR65 pKa = 11.84VIVIRR70 pKa = 11.84HH71 pKa = 4.59NVKK74 pKa = 10.47VIEE77 pKa = 4.53GQAVVLNGTQYY88 pKa = 11.37DD89 pKa = 3.79IVRR92 pKa = 11.84VSSNEE97 pKa = 3.55NFGLNRR103 pKa = 11.84YY104 pKa = 9.75DD105 pKa = 3.94YY106 pKa = 10.41LTLKK110 pKa = 9.73KK111 pKa = 9.68HH112 pKa = 5.98KK113 pKa = 10.48KK114 pKa = 9.32IGG116 pKa = 3.46
Molecular weight: 13.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10387 |
39 |
1603 |
230.8 |
26.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.633 ± 0.509 | 0.568 ± 0.127 |
6.412 ± 0.279 | 6.826 ± 0.531 |
4.246 ± 0.197 | 6.576 ± 0.471 |
1.54 ± 0.121 | 6.624 ± 0.211 |
8.636 ± 0.505 | 7.644 ± 0.389 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.628 ± 0.164 | 6.508 ± 0.274 |
2.821 ± 0.212 | 4.669 ± 0.24 |
4.169 ± 0.311 | 6.142 ± 0.284 |
6.094 ± 0.312 | 5.94 ± 0.204 |
1.454 ± 0.141 | 3.87 ± 0.283 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
