
Maize fine streak nucleorhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Nucleorhabdovirus
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
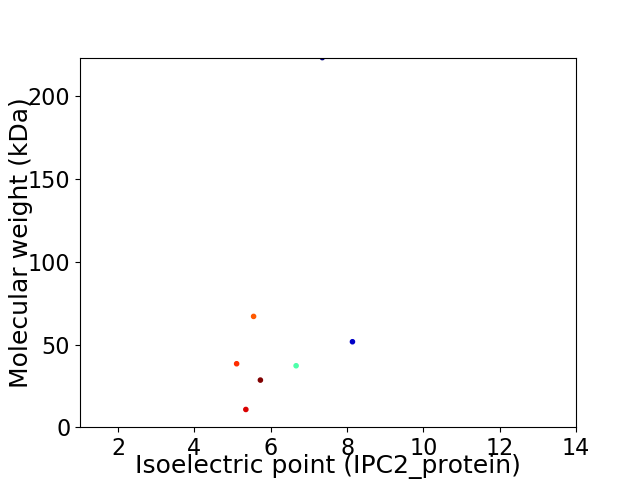
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q6E0X8|Q6E0X8_9RHAB N protein OS=Maize fine streak nucleorhabdovirus OX=209854 GN=N PE=4 SV=1
MM1 pKa = 7.08SQRR4 pKa = 11.84TLRR7 pKa = 11.84STRR10 pKa = 11.84SSQSKK15 pKa = 10.3KK16 pKa = 10.43GFANLEE22 pKa = 4.18SYY24 pKa = 10.32RR25 pKa = 11.84EE26 pKa = 3.99KK27 pKa = 10.88SVTEE31 pKa = 3.75ADD33 pKa = 3.39PAYY36 pKa = 10.8SSVQIPHH43 pKa = 6.38SVTEE47 pKa = 4.0STRR50 pKa = 11.84YY51 pKa = 9.9KK52 pKa = 10.73SLEE55 pKa = 4.07QVDD58 pKa = 4.46KK59 pKa = 11.34LPLSSDD65 pKa = 2.52IPAQIEE71 pKa = 4.63SQVEE75 pKa = 3.84MAPILTPEE83 pKa = 4.23RR84 pKa = 11.84KK85 pKa = 8.24EE86 pKa = 4.16TMKK89 pKa = 11.17EE90 pKa = 3.95EE91 pKa = 3.71ITKK94 pKa = 10.25FLMARR99 pKa = 11.84GCVPDD104 pKa = 3.93NMAMEE109 pKa = 4.54QLLKK113 pKa = 10.64AHH115 pKa = 6.58EE116 pKa = 4.69PIIDD120 pKa = 3.79NPAADD125 pKa = 5.1IIPDD129 pKa = 3.77TTYY132 pKa = 11.44LSAFCQGWTFCMRR145 pKa = 11.84VYY147 pKa = 11.41SDD149 pKa = 3.28MALNNLKK156 pKa = 8.87KK157 pKa = 9.11TIEE160 pKa = 4.86PIMSTLAATTNTMQGLTGEE179 pKa = 4.66LNHH182 pKa = 6.46TSKK185 pKa = 10.99LLKK188 pKa = 10.32LRR190 pKa = 11.84VSAVPFKK197 pKa = 10.59RR198 pKa = 11.84QSPKK202 pKa = 10.86DD203 pKa = 3.27VAARR207 pKa = 11.84INKK210 pKa = 7.43MKK212 pKa = 10.57QKK214 pKa = 10.12GRR216 pKa = 11.84RR217 pKa = 11.84AEE219 pKa = 5.14DD220 pKa = 2.78IMDD223 pKa = 4.14PMKK226 pKa = 10.79QPMVEE231 pKa = 4.01EE232 pKa = 4.54EE233 pKa = 3.9EE234 pKa = 5.72DD235 pKa = 3.3IDD237 pKa = 4.69EE238 pKa = 4.48EE239 pKa = 4.87SEE241 pKa = 4.65DD242 pKa = 3.78VFPEE246 pKa = 4.14YY247 pKa = 10.57SSSLLRR253 pKa = 11.84TTWDD257 pKa = 3.39EE258 pKa = 3.87MSDD261 pKa = 3.59DD262 pKa = 4.3LKK264 pKa = 11.21RR265 pKa = 11.84SLLYY269 pKa = 10.78GIIKK273 pKa = 10.18AAMDD277 pKa = 3.52IEE279 pKa = 4.77DD280 pKa = 3.98VGEE283 pKa = 4.28LDD285 pKa = 3.92SDD287 pKa = 3.85TMNTIEE293 pKa = 4.82RR294 pKa = 11.84SLKK297 pKa = 10.29GSQLTTAYY305 pKa = 10.73LMKK308 pKa = 10.73DD309 pKa = 3.11EE310 pKa = 5.1GVDD313 pKa = 3.07RR314 pKa = 11.84DD315 pKa = 3.68KK316 pKa = 10.94IRR318 pKa = 11.84RR319 pKa = 11.84YY320 pKa = 9.22IKK322 pKa = 10.16KK323 pKa = 8.87IWEE326 pKa = 3.95QAGLPNKK333 pKa = 9.88MEE335 pKa = 4.73IGDD338 pKa = 3.74
MM1 pKa = 7.08SQRR4 pKa = 11.84TLRR7 pKa = 11.84STRR10 pKa = 11.84SSQSKK15 pKa = 10.3KK16 pKa = 10.43GFANLEE22 pKa = 4.18SYY24 pKa = 10.32RR25 pKa = 11.84EE26 pKa = 3.99KK27 pKa = 10.88SVTEE31 pKa = 3.75ADD33 pKa = 3.39PAYY36 pKa = 10.8SSVQIPHH43 pKa = 6.38SVTEE47 pKa = 4.0STRR50 pKa = 11.84YY51 pKa = 9.9KK52 pKa = 10.73SLEE55 pKa = 4.07QVDD58 pKa = 4.46KK59 pKa = 11.34LPLSSDD65 pKa = 2.52IPAQIEE71 pKa = 4.63SQVEE75 pKa = 3.84MAPILTPEE83 pKa = 4.23RR84 pKa = 11.84KK85 pKa = 8.24EE86 pKa = 4.16TMKK89 pKa = 11.17EE90 pKa = 3.95EE91 pKa = 3.71ITKK94 pKa = 10.25FLMARR99 pKa = 11.84GCVPDD104 pKa = 3.93NMAMEE109 pKa = 4.54QLLKK113 pKa = 10.64AHH115 pKa = 6.58EE116 pKa = 4.69PIIDD120 pKa = 3.79NPAADD125 pKa = 5.1IIPDD129 pKa = 3.77TTYY132 pKa = 11.44LSAFCQGWTFCMRR145 pKa = 11.84VYY147 pKa = 11.41SDD149 pKa = 3.28MALNNLKK156 pKa = 8.87KK157 pKa = 9.11TIEE160 pKa = 4.86PIMSTLAATTNTMQGLTGEE179 pKa = 4.66LNHH182 pKa = 6.46TSKK185 pKa = 10.99LLKK188 pKa = 10.32LRR190 pKa = 11.84VSAVPFKK197 pKa = 10.59RR198 pKa = 11.84QSPKK202 pKa = 10.86DD203 pKa = 3.27VAARR207 pKa = 11.84INKK210 pKa = 7.43MKK212 pKa = 10.57QKK214 pKa = 10.12GRR216 pKa = 11.84RR217 pKa = 11.84AEE219 pKa = 5.14DD220 pKa = 2.78IMDD223 pKa = 4.14PMKK226 pKa = 10.79QPMVEE231 pKa = 4.01EE232 pKa = 4.54EE233 pKa = 3.9EE234 pKa = 5.72DD235 pKa = 3.3IDD237 pKa = 4.69EE238 pKa = 4.48EE239 pKa = 4.87SEE241 pKa = 4.65DD242 pKa = 3.78VFPEE246 pKa = 4.14YY247 pKa = 10.57SSSLLRR253 pKa = 11.84TTWDD257 pKa = 3.39EE258 pKa = 3.87MSDD261 pKa = 3.59DD262 pKa = 4.3LKK264 pKa = 11.21RR265 pKa = 11.84SLLYY269 pKa = 10.78GIIKK273 pKa = 10.18AAMDD277 pKa = 3.52IEE279 pKa = 4.77DD280 pKa = 3.98VGEE283 pKa = 4.28LDD285 pKa = 3.92SDD287 pKa = 3.85TMNTIEE293 pKa = 4.82RR294 pKa = 11.84SLKK297 pKa = 10.29GSQLTTAYY305 pKa = 10.73LMKK308 pKa = 10.73DD309 pKa = 3.11EE310 pKa = 5.1GVDD313 pKa = 3.07RR314 pKa = 11.84DD315 pKa = 3.68KK316 pKa = 10.94IRR318 pKa = 11.84RR319 pKa = 11.84YY320 pKa = 9.22IKK322 pKa = 10.16KK323 pKa = 8.87IWEE326 pKa = 3.95QAGLPNKK333 pKa = 9.88MEE335 pKa = 4.73IGDD338 pKa = 3.74
Molecular weight: 38.37 kDa
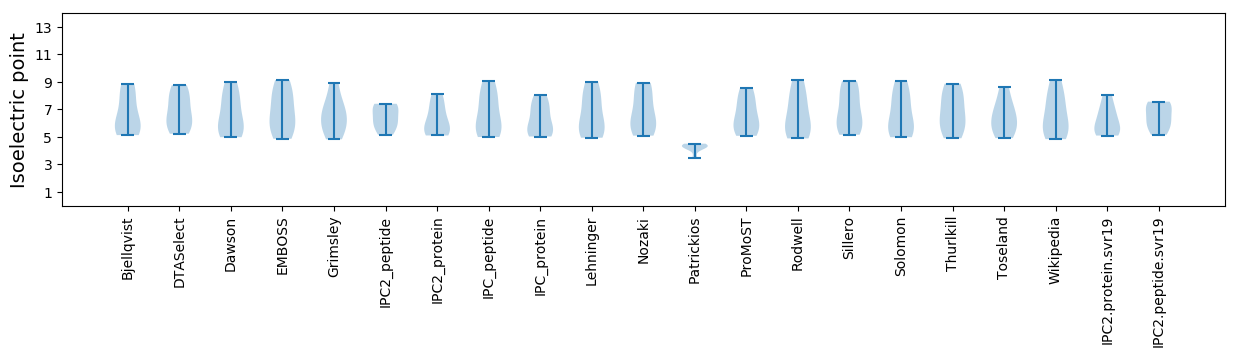
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6E0X8|Q6E0X8_9RHAB N protein OS=Maize fine streak nucleorhabdovirus OX=209854 GN=N PE=4 SV=1
MM1 pKa = 7.66NYY3 pKa = 9.79NRR5 pKa = 11.84LKK7 pKa = 11.04FEE9 pKa = 4.77DD10 pKa = 4.07AFLTPSADD18 pKa = 3.19YY19 pKa = 10.99AAFKK23 pKa = 10.51TPLPYY28 pKa = 9.76PDD30 pKa = 4.54LTTGPTEE37 pKa = 3.89VPYY40 pKa = 10.52TKK42 pKa = 10.33EE43 pKa = 3.78KK44 pKa = 10.71LITASYY50 pKa = 8.98PYY52 pKa = 8.9WTMTKK57 pKa = 10.2VNSGDD62 pKa = 3.06IKK64 pKa = 10.17TLGKK68 pKa = 8.51TVLTKK73 pKa = 11.04LSDD76 pKa = 3.39KK77 pKa = 10.92KK78 pKa = 10.02ITEE81 pKa = 3.79KK82 pKa = 10.44TLFDD86 pKa = 3.66MCRR89 pKa = 11.84IALSLKK95 pKa = 10.21APNGQDD101 pKa = 2.99VFKK104 pKa = 10.83IPTVTFAGAKK114 pKa = 9.34TGGQDD119 pKa = 3.44DD120 pKa = 4.55TNLVPVPSDD129 pKa = 3.16TSKK132 pKa = 7.65TTEE135 pKa = 3.84YY136 pKa = 10.48LQTVTSKK143 pKa = 11.03SSTSQATQKK152 pKa = 10.78KK153 pKa = 9.49SLSTYY158 pKa = 10.01VDD160 pKa = 2.84NDD162 pKa = 3.41ANYY165 pKa = 7.27EE166 pKa = 4.02TAIPYY171 pKa = 7.35FLCSFLRR178 pKa = 11.84LIVKK182 pKa = 9.91QPEE185 pKa = 3.62SWRR188 pKa = 11.84RR189 pKa = 11.84AFGDD193 pKa = 3.13IKK195 pKa = 10.2EE196 pKa = 4.37QYY198 pKa = 10.2GKK200 pKa = 10.83FYY202 pKa = 11.1GKK204 pKa = 8.98TSNLITNAEE213 pKa = 4.34DD214 pKa = 3.87DD215 pKa = 4.18LSIASVVKK223 pKa = 10.52VAFDD227 pKa = 3.4TFKK230 pKa = 10.89PIVNFIAYY238 pKa = 9.19VAGEE242 pKa = 4.2TDD244 pKa = 4.19KK245 pKa = 11.55GLSPNTKK252 pKa = 7.47EE253 pKa = 4.59HH254 pKa = 6.57GMFTYY259 pKa = 10.16FVGQHH264 pKa = 5.71LSFVGMHH271 pKa = 6.5VYY273 pKa = 10.67PMVAEE278 pKa = 4.84LMQKK282 pKa = 10.37CKK284 pKa = 10.62GIKK287 pKa = 9.23QDD289 pKa = 4.26LFLTFLDD296 pKa = 3.33VDD298 pKa = 3.84EE299 pKa = 4.97TKK301 pKa = 10.91EE302 pKa = 4.2SVKK305 pKa = 10.2EE306 pKa = 3.52ICRR309 pKa = 11.84IMTTYY314 pKa = 10.61DD315 pKa = 3.24APSQTDD321 pKa = 3.12RR322 pKa = 11.84ASRR325 pKa = 11.84DD326 pKa = 3.71FLWKK330 pKa = 8.15YY331 pKa = 10.84ARR333 pKa = 11.84VIDD336 pKa = 3.78EE337 pKa = 4.53GFFLKK342 pKa = 10.67LQNKK346 pKa = 8.46KK347 pKa = 10.4NKK349 pKa = 9.34EE350 pKa = 3.91FLYY353 pKa = 10.85GLALIHH359 pKa = 6.51EE360 pKa = 4.65KK361 pKa = 9.91MGLVRR366 pKa = 11.84SVSYY370 pKa = 11.05AKK372 pKa = 8.14PTNMAILQNQDD383 pKa = 2.72GLRR386 pKa = 11.84ADD388 pKa = 3.58AEE390 pKa = 4.44EE391 pKa = 4.12YY392 pKa = 11.32ANLFVFLYY400 pKa = 10.55KK401 pKa = 10.49KK402 pKa = 10.12ATGQKK407 pKa = 9.62EE408 pKa = 4.3GASPMDD414 pKa = 3.42QLRR417 pKa = 11.84SQLAGEE423 pKa = 4.44RR424 pKa = 11.84VTATIAEE431 pKa = 4.08ASAPKK436 pKa = 10.06RR437 pKa = 11.84SSDD440 pKa = 3.16GTGNVSKK447 pKa = 10.75KK448 pKa = 10.14KK449 pKa = 10.08SRR451 pKa = 11.84KK452 pKa = 9.24DD453 pKa = 3.24DD454 pKa = 3.33TGIASMEE461 pKa = 4.07FF462 pKa = 3.31
MM1 pKa = 7.66NYY3 pKa = 9.79NRR5 pKa = 11.84LKK7 pKa = 11.04FEE9 pKa = 4.77DD10 pKa = 4.07AFLTPSADD18 pKa = 3.19YY19 pKa = 10.99AAFKK23 pKa = 10.51TPLPYY28 pKa = 9.76PDD30 pKa = 4.54LTTGPTEE37 pKa = 3.89VPYY40 pKa = 10.52TKK42 pKa = 10.33EE43 pKa = 3.78KK44 pKa = 10.71LITASYY50 pKa = 8.98PYY52 pKa = 8.9WTMTKK57 pKa = 10.2VNSGDD62 pKa = 3.06IKK64 pKa = 10.17TLGKK68 pKa = 8.51TVLTKK73 pKa = 11.04LSDD76 pKa = 3.39KK77 pKa = 10.92KK78 pKa = 10.02ITEE81 pKa = 3.79KK82 pKa = 10.44TLFDD86 pKa = 3.66MCRR89 pKa = 11.84IALSLKK95 pKa = 10.21APNGQDD101 pKa = 2.99VFKK104 pKa = 10.83IPTVTFAGAKK114 pKa = 9.34TGGQDD119 pKa = 3.44DD120 pKa = 4.55TNLVPVPSDD129 pKa = 3.16TSKK132 pKa = 7.65TTEE135 pKa = 3.84YY136 pKa = 10.48LQTVTSKK143 pKa = 11.03SSTSQATQKK152 pKa = 10.78KK153 pKa = 9.49SLSTYY158 pKa = 10.01VDD160 pKa = 2.84NDD162 pKa = 3.41ANYY165 pKa = 7.27EE166 pKa = 4.02TAIPYY171 pKa = 7.35FLCSFLRR178 pKa = 11.84LIVKK182 pKa = 9.91QPEE185 pKa = 3.62SWRR188 pKa = 11.84RR189 pKa = 11.84AFGDD193 pKa = 3.13IKK195 pKa = 10.2EE196 pKa = 4.37QYY198 pKa = 10.2GKK200 pKa = 10.83FYY202 pKa = 11.1GKK204 pKa = 8.98TSNLITNAEE213 pKa = 4.34DD214 pKa = 3.87DD215 pKa = 4.18LSIASVVKK223 pKa = 10.52VAFDD227 pKa = 3.4TFKK230 pKa = 10.89PIVNFIAYY238 pKa = 9.19VAGEE242 pKa = 4.2TDD244 pKa = 4.19KK245 pKa = 11.55GLSPNTKK252 pKa = 7.47EE253 pKa = 4.59HH254 pKa = 6.57GMFTYY259 pKa = 10.16FVGQHH264 pKa = 5.71LSFVGMHH271 pKa = 6.5VYY273 pKa = 10.67PMVAEE278 pKa = 4.84LMQKK282 pKa = 10.37CKK284 pKa = 10.62GIKK287 pKa = 9.23QDD289 pKa = 4.26LFLTFLDD296 pKa = 3.33VDD298 pKa = 3.84EE299 pKa = 4.97TKK301 pKa = 10.91EE302 pKa = 4.2SVKK305 pKa = 10.2EE306 pKa = 3.52ICRR309 pKa = 11.84IMTTYY314 pKa = 10.61DD315 pKa = 3.24APSQTDD321 pKa = 3.12RR322 pKa = 11.84ASRR325 pKa = 11.84DD326 pKa = 3.71FLWKK330 pKa = 8.15YY331 pKa = 10.84ARR333 pKa = 11.84VIDD336 pKa = 3.78EE337 pKa = 4.53GFFLKK342 pKa = 10.67LQNKK346 pKa = 8.46KK347 pKa = 10.4NKK349 pKa = 9.34EE350 pKa = 3.91FLYY353 pKa = 10.85GLALIHH359 pKa = 6.51EE360 pKa = 4.65KK361 pKa = 9.91MGLVRR366 pKa = 11.84SVSYY370 pKa = 11.05AKK372 pKa = 8.14PTNMAILQNQDD383 pKa = 2.72GLRR386 pKa = 11.84ADD388 pKa = 3.58AEE390 pKa = 4.44EE391 pKa = 4.12YY392 pKa = 11.32ANLFVFLYY400 pKa = 10.55KK401 pKa = 10.49KK402 pKa = 10.12ATGQKK407 pKa = 9.62EE408 pKa = 4.3GASPMDD414 pKa = 3.42QLRR417 pKa = 11.84SQLAGEE423 pKa = 4.44RR424 pKa = 11.84VTATIAEE431 pKa = 4.08ASAPKK436 pKa = 10.06RR437 pKa = 11.84SSDD440 pKa = 3.16GTGNVSKK447 pKa = 10.75KK448 pKa = 10.14KK449 pKa = 10.08SRR451 pKa = 11.84KK452 pKa = 9.24DD453 pKa = 3.24DD454 pKa = 3.33TGIASMEE461 pKa = 4.07FF462 pKa = 3.31
Molecular weight: 51.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4006 |
93 |
1944 |
572.3 |
65.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.818 ± 0.583 | 1.523 ± 0.285 |
5.891 ± 0.442 | 6.166 ± 0.42 |
3.819 ± 0.458 | 5.117 ± 0.259 |
2.122 ± 0.279 | 7.639 ± 0.641 |
6.965 ± 0.574 | 9.411 ± 0.821 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.12 ± 0.316 | 4.269 ± 0.454 |
4.169 ± 0.171 | 3.27 ± 0.291 |
4.868 ± 0.32 | 8.462 ± 0.388 |
6.99 ± 0.827 | 5.542 ± 0.396 |
1.498 ± 0.234 | 4.343 ± 0.251 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
