
Psychromonas sp. B3M02
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Psychromonadaceae; Psychromonas; unclassified Psychromonas
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
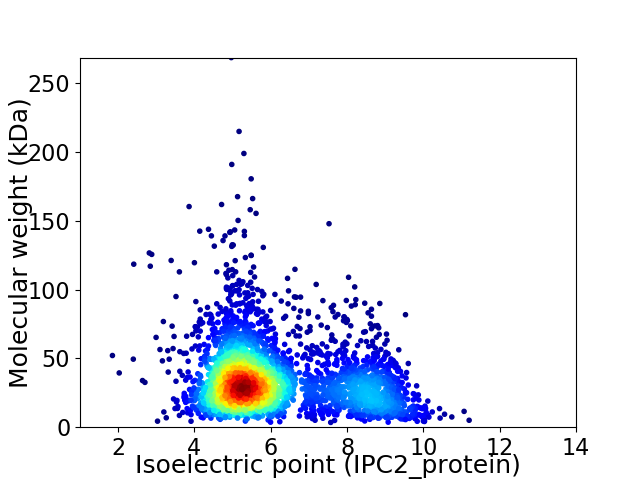
Virtual 2D-PAGE plot for 3376 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
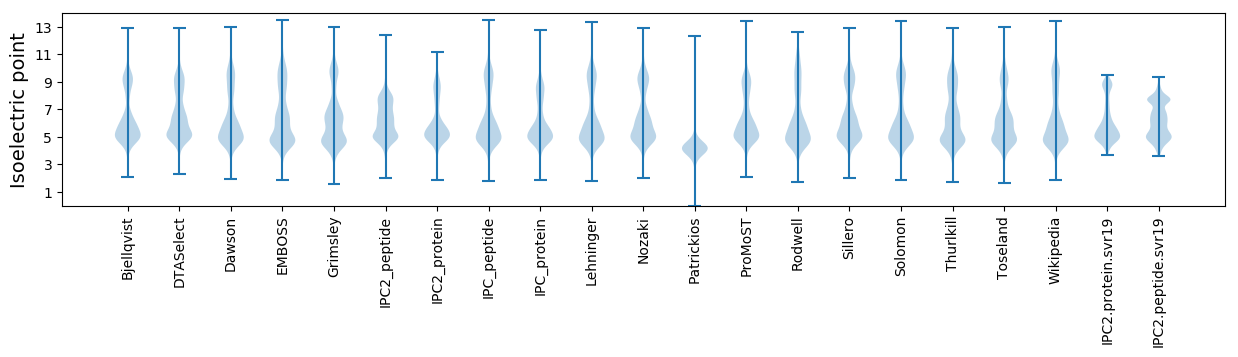
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A366WD60|A0A366WD60_9GAMM Exodeoxyribonuclease III OS=Psychromonas sp. B3M02 OX=2267226 GN=DS885_00805 PE=3 SV=1
MM1 pKa = 7.45LLIAGYY7 pKa = 9.87LLPIAVNASNLDD19 pKa = 3.83LATKK23 pKa = 9.96PLYY26 pKa = 10.16TGTSEE31 pKa = 4.52APMMMLTMGRR41 pKa = 11.84DD42 pKa = 2.96HH43 pKa = 6.59TLFYY47 pKa = 10.59EE48 pKa = 4.94AYY50 pKa = 10.01NDD52 pKa = 4.77ASDD55 pKa = 4.66LDD57 pKa = 4.27GDD59 pKa = 4.63GEE61 pKa = 5.12LDD63 pKa = 3.48TTFKK67 pKa = 10.67PDD69 pKa = 3.32YY70 pKa = 9.25TYY72 pKa = 11.23EE73 pKa = 4.5GYY75 pKa = 10.56FDD77 pKa = 3.64STACYY82 pKa = 9.92KK83 pKa = 10.46YY84 pKa = 10.33RR85 pKa = 11.84SNKK88 pKa = 9.68SRR90 pKa = 11.84FKK92 pKa = 10.44PKK94 pKa = 9.95TIEE97 pKa = 4.31TVCSNNNHH105 pKa = 5.99WSGNFLNYY113 pKa = 9.65LTMTRR118 pKa = 11.84MDD120 pKa = 3.85VLRR123 pKa = 11.84KK124 pKa = 9.6VLYY127 pKa = 10.55GGTRR131 pKa = 11.84STDD134 pKa = 3.59DD135 pKa = 3.43EE136 pKa = 5.34DD137 pKa = 3.57EE138 pKa = 4.8TILEE142 pKa = 4.32RR143 pKa = 11.84VFIPQDD149 pKa = 2.91AHH151 pKa = 6.55SWAKK155 pKa = 10.39AYY157 pKa = 9.72TSVEE161 pKa = 3.54VDD163 pKa = 3.86GYY165 pKa = 11.09DD166 pKa = 3.12ISDD169 pKa = 3.66YY170 pKa = 10.97TPYY173 pKa = 11.04SLPDD177 pKa = 3.02SGKK180 pKa = 7.96QHH182 pKa = 6.61FFGTVTYY189 pKa = 10.33SGLGEE194 pKa = 4.27DD195 pKa = 3.92PILRR199 pKa = 11.84VRR201 pKa = 11.84EE202 pKa = 4.14NISYY206 pKa = 10.89SSDD209 pKa = 3.22DD210 pKa = 3.18WGVWSWASTEE220 pKa = 4.27RR221 pKa = 11.84PVLDD225 pKa = 4.32ASGDD229 pKa = 3.79DD230 pKa = 3.35YY231 pKa = 11.61VVRR234 pKa = 11.84VEE236 pKa = 4.02ACVDD240 pKa = 3.56GFTDD244 pKa = 5.16GNNCDD249 pKa = 4.87LYY251 pKa = 10.7PSGNYY256 pKa = 9.47KK257 pKa = 8.81PTGLLHH263 pKa = 7.69DD264 pKa = 4.59YY265 pKa = 11.58GEE267 pKa = 4.38TDD269 pKa = 3.12QMLFGLLTGSYY280 pKa = 10.35NKK282 pKa = 9.97NLSGGVLRR290 pKa = 11.84RR291 pKa = 11.84AISEE295 pKa = 3.89FSEE298 pKa = 4.57EE299 pKa = 4.05VDD301 pKa = 3.11SDD303 pKa = 3.64TGQFTSTEE311 pKa = 4.07DD312 pKa = 4.05GIVATIDD319 pKa = 3.08KK320 pKa = 10.69LRR322 pKa = 11.84IHH324 pKa = 7.1GFNYY328 pKa = 10.2SNKK331 pKa = 9.49TYY333 pKa = 9.63ATNCGWVVNGAISDD347 pKa = 4.11GQCASWGNPAGEE359 pKa = 4.16MLYY362 pKa = 10.76EE363 pKa = 4.12AMRR366 pKa = 11.84YY367 pKa = 8.7YY368 pKa = 10.84SGVEE372 pKa = 4.05SASPSYY378 pKa = 9.71ATSGSYY384 pKa = 10.16DD385 pKa = 3.57AEE387 pKa = 4.44VGLTSPDD394 pKa = 2.98WDD396 pKa = 4.68DD397 pKa = 3.81PFEE400 pKa = 5.48DD401 pKa = 4.88RR402 pKa = 11.84GTCANPNVLLISDD415 pKa = 4.59ISPSYY420 pKa = 11.17DD421 pKa = 3.22SDD423 pKa = 3.68QLPGVYY429 pKa = 9.94SEE431 pKa = 4.58FTKK434 pKa = 10.82SANNDD439 pKa = 2.9EE440 pKa = 5.37AYY442 pKa = 10.44DD443 pKa = 3.89GTTLPGFSVSEE454 pKa = 3.9LLEE457 pKa = 4.51TISTSEE463 pKa = 4.34GKK465 pKa = 8.93TSGQFFIGQSGTDD478 pKa = 3.24NDD480 pKa = 4.48GAPTAKK486 pKa = 10.43DD487 pKa = 3.04ITGLEE492 pKa = 4.4SVRR495 pKa = 11.84GLAPAEE501 pKa = 3.98PTKK504 pKa = 10.68KK505 pKa = 10.45GSYY508 pKa = 9.86SSAAVAYY515 pKa = 9.07YY516 pKa = 8.81GHH518 pKa = 7.27RR519 pKa = 11.84NDD521 pKa = 3.03IHH523 pKa = 5.61LTKK526 pKa = 10.95DD527 pKa = 2.61EE528 pKa = 4.5DD529 pKa = 4.0QNVTTMVVALASTLPEE545 pKa = 4.17IEE547 pKa = 3.98VDD549 pKa = 3.29VDD551 pKa = 3.84GEE553 pKa = 4.44VIKK556 pKa = 10.51IVPFAKK562 pKa = 10.2SVGWGNKK569 pKa = 8.22ISADD573 pKa = 3.49EE574 pKa = 4.98GDD576 pKa = 4.08FQPTNSIIDD585 pKa = 3.56WYY587 pKa = 10.26IEE589 pKa = 4.34SITDD593 pKa = 3.27TKK595 pKa = 11.23GVFRR599 pKa = 11.84INYY602 pKa = 8.76EE603 pKa = 4.03DD604 pKa = 3.82VEE606 pKa = 4.42QGADD610 pKa = 3.14HH611 pKa = 7.22DD612 pKa = 3.7MDD614 pKa = 4.61MIVKK618 pKa = 8.05YY619 pKa = 9.03TYY621 pKa = 9.01EE622 pKa = 4.08VKK624 pKa = 10.66EE625 pKa = 4.49GLCKK629 pKa = 10.18TYY631 pKa = 10.9DD632 pKa = 3.43SDD634 pKa = 4.76GDD636 pKa = 3.91CTEE639 pKa = 3.95TAKK642 pKa = 10.84GVEE645 pKa = 4.0ITLDD649 pKa = 3.23STYY652 pKa = 11.27AAGGVDD658 pKa = 3.06QHH660 pKa = 7.59AGYY663 pKa = 9.94IISGTTNDD671 pKa = 3.45GMYY674 pKa = 10.77LEE676 pKa = 4.83VKK678 pKa = 10.06DD679 pKa = 4.47VSGANIKK686 pKa = 10.67YY687 pKa = 10.64YY688 pKa = 10.87LDD690 pKa = 4.02TPDD693 pKa = 3.09STVYY697 pKa = 10.87DD698 pKa = 3.56NRR700 pKa = 11.84DD701 pKa = 3.03ISGSTSNLGLYY712 pKa = 7.44ATRR715 pKa = 11.84NFFPSDD721 pKa = 2.95EE722 pKa = 4.33SAADD726 pKa = 3.89FLPSPLWYY734 pKa = 9.32AAKK737 pKa = 9.96WGGFTDD743 pKa = 3.53SDD745 pKa = 4.4GDD747 pKa = 3.82GTPNLDD753 pKa = 4.55SEE755 pKa = 4.7WDD757 pKa = 3.39TSGADD762 pKa = 4.07DD763 pKa = 4.97GGADD767 pKa = 4.45GDD769 pKa = 4.06PDD771 pKa = 3.65NYY773 pKa = 10.67FPVTNAGEE781 pKa = 4.66LKK783 pKa = 10.51DD784 pKa = 3.73QLAAAFDD791 pKa = 3.92LAYY794 pKa = 10.72DD795 pKa = 4.05GLLTGASATYY805 pKa = 10.45SSNLLSSGSLAYY817 pKa = 10.15QSSFEE822 pKa = 4.1EE823 pKa = 4.54EE824 pKa = 3.99TWNGDD829 pKa = 3.07VNAYY833 pKa = 9.03QVSSDD838 pKa = 3.73GEE840 pKa = 4.41YY841 pKa = 10.95ASNATWSAATQLDD854 pKa = 4.29AMDD857 pKa = 5.26PDD859 pKa = 3.43NRR861 pKa = 11.84HH862 pKa = 5.95IYY864 pKa = 8.58TRR866 pKa = 11.84SNEE869 pKa = 3.9SSLLGSQGAVFEE881 pKa = 4.61FTTSAISTTLLSSLTDD897 pKa = 3.28SLTSVSTSTLTDD909 pKa = 3.36SFSEE913 pKa = 4.22SQLFSLLGDD922 pKa = 3.63VTGSTTEE929 pKa = 3.69KK930 pKa = 10.43LAYY933 pKa = 7.68LTNVVGYY940 pKa = 10.31IRR942 pKa = 11.84GDD944 pKa = 3.25RR945 pKa = 11.84DD946 pKa = 3.58NEE948 pKa = 4.06VTTVVDD954 pKa = 4.14GFRR957 pKa = 11.84EE958 pKa = 4.03RR959 pKa = 11.84SSVLGDD965 pKa = 3.34IVNASPYY972 pKa = 9.22YY973 pKa = 9.54VSSAKK978 pKa = 10.52DD979 pKa = 3.11HH980 pKa = 5.72TVATEE985 pKa = 4.14VVAFAANDD993 pKa = 3.77GMVHH997 pKa = 6.77IVDD1000 pKa = 4.21ADD1002 pKa = 3.86DD1003 pKa = 4.34GKK1005 pKa = 11.18EE1006 pKa = 3.59LAAYY1010 pKa = 8.88IPSGVYY1016 pKa = 9.81DD1017 pKa = 3.68ALGNLVDD1024 pKa = 3.36SAYY1027 pKa = 9.35IHH1029 pKa = 6.86EE1030 pKa = 4.28YY1031 pKa = 10.74SVDD1034 pKa = 3.5GGLNAYY1040 pKa = 10.3SDD1042 pKa = 4.44EE1043 pKa = 4.3NDD1045 pKa = 5.32LIDD1048 pKa = 3.6VDD1050 pKa = 4.03INGVSTEE1057 pKa = 4.05VARR1060 pKa = 11.84TTLVGRR1066 pKa = 11.84LGLGVKK1072 pKa = 9.81GLYY1075 pKa = 10.61AIDD1078 pKa = 3.87VSDD1081 pKa = 3.92ISNPTTDD1088 pKa = 2.88MMRR1091 pKa = 11.84WEE1093 pKa = 4.47ITTDD1097 pKa = 2.9TDD1099 pKa = 3.44GFEE1102 pKa = 4.36GLGLSVSEE1110 pKa = 4.02PTIVEE1115 pKa = 4.2LANGKK1120 pKa = 10.33ASVVFANGYY1129 pKa = 10.34NSTDD1133 pKa = 3.61DD1134 pKa = 3.44EE1135 pKa = 4.55GAIYY1139 pKa = 10.2IADD1142 pKa = 4.0LTDD1145 pKa = 3.4GSLIKK1150 pKa = 10.53RR1151 pKa = 11.84LTVGVQTDD1159 pKa = 3.59PTGEE1163 pKa = 3.99DD1164 pKa = 3.31RR1165 pKa = 11.84PNALAQPLLVDD1176 pKa = 4.01NNSDD1180 pKa = 3.93GIADD1184 pKa = 3.69YY1185 pKa = 10.9LYY1187 pKa = 11.04AGDD1190 pKa = 4.98LFGNMWAFDD1199 pKa = 3.74ISSEE1203 pKa = 4.74DD1204 pKa = 3.54PSEE1207 pKa = 3.82WGIKK1211 pKa = 8.01KK1212 pKa = 7.94TAGAPLFTALSPTLDD1227 pKa = 3.43NQEE1230 pKa = 4.47YY1231 pKa = 8.23MSQPITSRR1239 pKa = 11.84PSVVRR1244 pKa = 11.84HH1245 pKa = 5.99PDD1247 pKa = 3.33GEE1249 pKa = 4.55GVLIAFGTGKK1259 pKa = 10.38YY1260 pKa = 9.73VEE1262 pKa = 4.92SDD1264 pKa = 3.3DD1265 pKa = 5.14TSADD1269 pKa = 3.68DD1270 pKa = 3.85QATQSFYY1277 pKa = 11.39VVEE1280 pKa = 4.95DD1281 pKa = 3.71NLGSAVVTAVRR1292 pKa = 11.84SSSGFSNLQKK1302 pKa = 10.8QSITKK1307 pKa = 9.63EE1308 pKa = 3.87SSTNRR1313 pKa = 11.84LLSSNSLNWDD1323 pKa = 4.05EE1324 pKa = 5.27ISGFYY1329 pKa = 9.83IDD1331 pKa = 5.78LVNTEE1336 pKa = 5.06DD1337 pKa = 4.87GNTNNYY1343 pKa = 9.89GEE1345 pKa = 4.32RR1346 pKa = 11.84QVVASTVLGTKK1357 pKa = 10.06VSFVTLIPGTDD1368 pKa = 3.2PCVAGGTGWYY1378 pKa = 8.87MEE1380 pKa = 4.58LNLYY1384 pKa = 8.27TGQTWDD1390 pKa = 3.05TGTEE1394 pKa = 4.1VEE1396 pKa = 5.18DD1397 pKa = 4.92DD1398 pKa = 3.72PSTTVDD1404 pKa = 3.41EE1405 pKa = 4.7SEE1407 pKa = 5.04EE1408 pKa = 4.13IPTDD1412 pKa = 3.42SSNQFLDD1419 pKa = 4.34GVATGLSSVASVTTSTEE1436 pKa = 4.02TITNEE1441 pKa = 3.98DD1442 pKa = 3.6GTTEE1446 pKa = 3.97EE1447 pKa = 4.26VTVFEE1452 pKa = 4.4SAEE1455 pKa = 3.97GLTCITLSTNKK1466 pKa = 7.78QTCFEE1471 pKa = 5.31DD1472 pKa = 3.79DD1473 pKa = 4.11LNITGPVSWRR1483 pKa = 11.84QLYY1486 pKa = 9.99
MM1 pKa = 7.45LLIAGYY7 pKa = 9.87LLPIAVNASNLDD19 pKa = 3.83LATKK23 pKa = 9.96PLYY26 pKa = 10.16TGTSEE31 pKa = 4.52APMMMLTMGRR41 pKa = 11.84DD42 pKa = 2.96HH43 pKa = 6.59TLFYY47 pKa = 10.59EE48 pKa = 4.94AYY50 pKa = 10.01NDD52 pKa = 4.77ASDD55 pKa = 4.66LDD57 pKa = 4.27GDD59 pKa = 4.63GEE61 pKa = 5.12LDD63 pKa = 3.48TTFKK67 pKa = 10.67PDD69 pKa = 3.32YY70 pKa = 9.25TYY72 pKa = 11.23EE73 pKa = 4.5GYY75 pKa = 10.56FDD77 pKa = 3.64STACYY82 pKa = 9.92KK83 pKa = 10.46YY84 pKa = 10.33RR85 pKa = 11.84SNKK88 pKa = 9.68SRR90 pKa = 11.84FKK92 pKa = 10.44PKK94 pKa = 9.95TIEE97 pKa = 4.31TVCSNNNHH105 pKa = 5.99WSGNFLNYY113 pKa = 9.65LTMTRR118 pKa = 11.84MDD120 pKa = 3.85VLRR123 pKa = 11.84KK124 pKa = 9.6VLYY127 pKa = 10.55GGTRR131 pKa = 11.84STDD134 pKa = 3.59DD135 pKa = 3.43EE136 pKa = 5.34DD137 pKa = 3.57EE138 pKa = 4.8TILEE142 pKa = 4.32RR143 pKa = 11.84VFIPQDD149 pKa = 2.91AHH151 pKa = 6.55SWAKK155 pKa = 10.39AYY157 pKa = 9.72TSVEE161 pKa = 3.54VDD163 pKa = 3.86GYY165 pKa = 11.09DD166 pKa = 3.12ISDD169 pKa = 3.66YY170 pKa = 10.97TPYY173 pKa = 11.04SLPDD177 pKa = 3.02SGKK180 pKa = 7.96QHH182 pKa = 6.61FFGTVTYY189 pKa = 10.33SGLGEE194 pKa = 4.27DD195 pKa = 3.92PILRR199 pKa = 11.84VRR201 pKa = 11.84EE202 pKa = 4.14NISYY206 pKa = 10.89SSDD209 pKa = 3.22DD210 pKa = 3.18WGVWSWASTEE220 pKa = 4.27RR221 pKa = 11.84PVLDD225 pKa = 4.32ASGDD229 pKa = 3.79DD230 pKa = 3.35YY231 pKa = 11.61VVRR234 pKa = 11.84VEE236 pKa = 4.02ACVDD240 pKa = 3.56GFTDD244 pKa = 5.16GNNCDD249 pKa = 4.87LYY251 pKa = 10.7PSGNYY256 pKa = 9.47KK257 pKa = 8.81PTGLLHH263 pKa = 7.69DD264 pKa = 4.59YY265 pKa = 11.58GEE267 pKa = 4.38TDD269 pKa = 3.12QMLFGLLTGSYY280 pKa = 10.35NKK282 pKa = 9.97NLSGGVLRR290 pKa = 11.84RR291 pKa = 11.84AISEE295 pKa = 3.89FSEE298 pKa = 4.57EE299 pKa = 4.05VDD301 pKa = 3.11SDD303 pKa = 3.64TGQFTSTEE311 pKa = 4.07DD312 pKa = 4.05GIVATIDD319 pKa = 3.08KK320 pKa = 10.69LRR322 pKa = 11.84IHH324 pKa = 7.1GFNYY328 pKa = 10.2SNKK331 pKa = 9.49TYY333 pKa = 9.63ATNCGWVVNGAISDD347 pKa = 4.11GQCASWGNPAGEE359 pKa = 4.16MLYY362 pKa = 10.76EE363 pKa = 4.12AMRR366 pKa = 11.84YY367 pKa = 8.7YY368 pKa = 10.84SGVEE372 pKa = 4.05SASPSYY378 pKa = 9.71ATSGSYY384 pKa = 10.16DD385 pKa = 3.57AEE387 pKa = 4.44VGLTSPDD394 pKa = 2.98WDD396 pKa = 4.68DD397 pKa = 3.81PFEE400 pKa = 5.48DD401 pKa = 4.88RR402 pKa = 11.84GTCANPNVLLISDD415 pKa = 4.59ISPSYY420 pKa = 11.17DD421 pKa = 3.22SDD423 pKa = 3.68QLPGVYY429 pKa = 9.94SEE431 pKa = 4.58FTKK434 pKa = 10.82SANNDD439 pKa = 2.9EE440 pKa = 5.37AYY442 pKa = 10.44DD443 pKa = 3.89GTTLPGFSVSEE454 pKa = 3.9LLEE457 pKa = 4.51TISTSEE463 pKa = 4.34GKK465 pKa = 8.93TSGQFFIGQSGTDD478 pKa = 3.24NDD480 pKa = 4.48GAPTAKK486 pKa = 10.43DD487 pKa = 3.04ITGLEE492 pKa = 4.4SVRR495 pKa = 11.84GLAPAEE501 pKa = 3.98PTKK504 pKa = 10.68KK505 pKa = 10.45GSYY508 pKa = 9.86SSAAVAYY515 pKa = 9.07YY516 pKa = 8.81GHH518 pKa = 7.27RR519 pKa = 11.84NDD521 pKa = 3.03IHH523 pKa = 5.61LTKK526 pKa = 10.95DD527 pKa = 2.61EE528 pKa = 4.5DD529 pKa = 4.0QNVTTMVVALASTLPEE545 pKa = 4.17IEE547 pKa = 3.98VDD549 pKa = 3.29VDD551 pKa = 3.84GEE553 pKa = 4.44VIKK556 pKa = 10.51IVPFAKK562 pKa = 10.2SVGWGNKK569 pKa = 8.22ISADD573 pKa = 3.49EE574 pKa = 4.98GDD576 pKa = 4.08FQPTNSIIDD585 pKa = 3.56WYY587 pKa = 10.26IEE589 pKa = 4.34SITDD593 pKa = 3.27TKK595 pKa = 11.23GVFRR599 pKa = 11.84INYY602 pKa = 8.76EE603 pKa = 4.03DD604 pKa = 3.82VEE606 pKa = 4.42QGADD610 pKa = 3.14HH611 pKa = 7.22DD612 pKa = 3.7MDD614 pKa = 4.61MIVKK618 pKa = 8.05YY619 pKa = 9.03TYY621 pKa = 9.01EE622 pKa = 4.08VKK624 pKa = 10.66EE625 pKa = 4.49GLCKK629 pKa = 10.18TYY631 pKa = 10.9DD632 pKa = 3.43SDD634 pKa = 4.76GDD636 pKa = 3.91CTEE639 pKa = 3.95TAKK642 pKa = 10.84GVEE645 pKa = 4.0ITLDD649 pKa = 3.23STYY652 pKa = 11.27AAGGVDD658 pKa = 3.06QHH660 pKa = 7.59AGYY663 pKa = 9.94IISGTTNDD671 pKa = 3.45GMYY674 pKa = 10.77LEE676 pKa = 4.83VKK678 pKa = 10.06DD679 pKa = 4.47VSGANIKK686 pKa = 10.67YY687 pKa = 10.64YY688 pKa = 10.87LDD690 pKa = 4.02TPDD693 pKa = 3.09STVYY697 pKa = 10.87DD698 pKa = 3.56NRR700 pKa = 11.84DD701 pKa = 3.03ISGSTSNLGLYY712 pKa = 7.44ATRR715 pKa = 11.84NFFPSDD721 pKa = 2.95EE722 pKa = 4.33SAADD726 pKa = 3.89FLPSPLWYY734 pKa = 9.32AAKK737 pKa = 9.96WGGFTDD743 pKa = 3.53SDD745 pKa = 4.4GDD747 pKa = 3.82GTPNLDD753 pKa = 4.55SEE755 pKa = 4.7WDD757 pKa = 3.39TSGADD762 pKa = 4.07DD763 pKa = 4.97GGADD767 pKa = 4.45GDD769 pKa = 4.06PDD771 pKa = 3.65NYY773 pKa = 10.67FPVTNAGEE781 pKa = 4.66LKK783 pKa = 10.51DD784 pKa = 3.73QLAAAFDD791 pKa = 3.92LAYY794 pKa = 10.72DD795 pKa = 4.05GLLTGASATYY805 pKa = 10.45SSNLLSSGSLAYY817 pKa = 10.15QSSFEE822 pKa = 4.1EE823 pKa = 4.54EE824 pKa = 3.99TWNGDD829 pKa = 3.07VNAYY833 pKa = 9.03QVSSDD838 pKa = 3.73GEE840 pKa = 4.41YY841 pKa = 10.95ASNATWSAATQLDD854 pKa = 4.29AMDD857 pKa = 5.26PDD859 pKa = 3.43NRR861 pKa = 11.84HH862 pKa = 5.95IYY864 pKa = 8.58TRR866 pKa = 11.84SNEE869 pKa = 3.9SSLLGSQGAVFEE881 pKa = 4.61FTTSAISTTLLSSLTDD897 pKa = 3.28SLTSVSTSTLTDD909 pKa = 3.36SFSEE913 pKa = 4.22SQLFSLLGDD922 pKa = 3.63VTGSTTEE929 pKa = 3.69KK930 pKa = 10.43LAYY933 pKa = 7.68LTNVVGYY940 pKa = 10.31IRR942 pKa = 11.84GDD944 pKa = 3.25RR945 pKa = 11.84DD946 pKa = 3.58NEE948 pKa = 4.06VTTVVDD954 pKa = 4.14GFRR957 pKa = 11.84EE958 pKa = 4.03RR959 pKa = 11.84SSVLGDD965 pKa = 3.34IVNASPYY972 pKa = 9.22YY973 pKa = 9.54VSSAKK978 pKa = 10.52DD979 pKa = 3.11HH980 pKa = 5.72TVATEE985 pKa = 4.14VVAFAANDD993 pKa = 3.77GMVHH997 pKa = 6.77IVDD1000 pKa = 4.21ADD1002 pKa = 3.86DD1003 pKa = 4.34GKK1005 pKa = 11.18EE1006 pKa = 3.59LAAYY1010 pKa = 8.88IPSGVYY1016 pKa = 9.81DD1017 pKa = 3.68ALGNLVDD1024 pKa = 3.36SAYY1027 pKa = 9.35IHH1029 pKa = 6.86EE1030 pKa = 4.28YY1031 pKa = 10.74SVDD1034 pKa = 3.5GGLNAYY1040 pKa = 10.3SDD1042 pKa = 4.44EE1043 pKa = 4.3NDD1045 pKa = 5.32LIDD1048 pKa = 3.6VDD1050 pKa = 4.03INGVSTEE1057 pKa = 4.05VARR1060 pKa = 11.84TTLVGRR1066 pKa = 11.84LGLGVKK1072 pKa = 9.81GLYY1075 pKa = 10.61AIDD1078 pKa = 3.87VSDD1081 pKa = 3.92ISNPTTDD1088 pKa = 2.88MMRR1091 pKa = 11.84WEE1093 pKa = 4.47ITTDD1097 pKa = 2.9TDD1099 pKa = 3.44GFEE1102 pKa = 4.36GLGLSVSEE1110 pKa = 4.02PTIVEE1115 pKa = 4.2LANGKK1120 pKa = 10.33ASVVFANGYY1129 pKa = 10.34NSTDD1133 pKa = 3.61DD1134 pKa = 3.44EE1135 pKa = 4.55GAIYY1139 pKa = 10.2IADD1142 pKa = 4.0LTDD1145 pKa = 3.4GSLIKK1150 pKa = 10.53RR1151 pKa = 11.84LTVGVQTDD1159 pKa = 3.59PTGEE1163 pKa = 3.99DD1164 pKa = 3.31RR1165 pKa = 11.84PNALAQPLLVDD1176 pKa = 4.01NNSDD1180 pKa = 3.93GIADD1184 pKa = 3.69YY1185 pKa = 10.9LYY1187 pKa = 11.04AGDD1190 pKa = 4.98LFGNMWAFDD1199 pKa = 3.74ISSEE1203 pKa = 4.74DD1204 pKa = 3.54PSEE1207 pKa = 3.82WGIKK1211 pKa = 8.01KK1212 pKa = 7.94TAGAPLFTALSPTLDD1227 pKa = 3.43NQEE1230 pKa = 4.47YY1231 pKa = 8.23MSQPITSRR1239 pKa = 11.84PSVVRR1244 pKa = 11.84HH1245 pKa = 5.99PDD1247 pKa = 3.33GEE1249 pKa = 4.55GVLIAFGTGKK1259 pKa = 10.38YY1260 pKa = 9.73VEE1262 pKa = 4.92SDD1264 pKa = 3.3DD1265 pKa = 5.14TSADD1269 pKa = 3.68DD1270 pKa = 3.85QATQSFYY1277 pKa = 11.39VVEE1280 pKa = 4.95DD1281 pKa = 3.71NLGSAVVTAVRR1292 pKa = 11.84SSSGFSNLQKK1302 pKa = 10.8QSITKK1307 pKa = 9.63EE1308 pKa = 3.87SSTNRR1313 pKa = 11.84LLSSNSLNWDD1323 pKa = 4.05EE1324 pKa = 5.27ISGFYY1329 pKa = 9.83IDD1331 pKa = 5.78LVNTEE1336 pKa = 5.06DD1337 pKa = 4.87GNTNNYY1343 pKa = 9.89GEE1345 pKa = 4.32RR1346 pKa = 11.84QVVASTVLGTKK1357 pKa = 10.06VSFVTLIPGTDD1368 pKa = 3.2PCVAGGTGWYY1378 pKa = 8.87MEE1380 pKa = 4.58LNLYY1384 pKa = 8.27TGQTWDD1390 pKa = 3.05TGTEE1394 pKa = 4.1VEE1396 pKa = 5.18DD1397 pKa = 4.92DD1398 pKa = 3.72PSTTVDD1404 pKa = 3.41EE1405 pKa = 4.7SEE1407 pKa = 5.04EE1408 pKa = 4.13IPTDD1412 pKa = 3.42SSNQFLDD1419 pKa = 4.34GVATGLSSVASVTTSTEE1436 pKa = 4.02TITNEE1441 pKa = 3.98DD1442 pKa = 3.6GTTEE1446 pKa = 3.97EE1447 pKa = 4.26VTVFEE1452 pKa = 4.4SAEE1455 pKa = 3.97GLTCITLSTNKK1466 pKa = 7.78QTCFEE1471 pKa = 5.31DD1472 pKa = 3.79DD1473 pKa = 4.11LNITGPVSWRR1483 pKa = 11.84QLYY1486 pKa = 9.99
Molecular weight: 160.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A366W235|A0A366W235_9GAMM DUF1488 domain-containing protein OS=Psychromonas sp. B3M02 OX=2267226 GN=DS885_11835 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNTKK11 pKa = 10.0RR12 pKa = 11.84KK13 pKa = 8.73RR14 pKa = 11.84SHH16 pKa = 6.16GFRR19 pKa = 11.84TRR21 pKa = 11.84MATKK25 pKa = 10.18NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.5GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNTKK11 pKa = 10.0RR12 pKa = 11.84KK13 pKa = 8.73RR14 pKa = 11.84SHH16 pKa = 6.16GFRR19 pKa = 11.84TRR21 pKa = 11.84MATKK25 pKa = 10.18NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.5GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
Molecular weight: 5.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1096779 |
31 |
2425 |
324.9 |
36.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.36 ± 0.043 | 1.041 ± 0.014 |
5.554 ± 0.039 | 6.112 ± 0.043 |
4.293 ± 0.031 | 6.418 ± 0.044 |
2.136 ± 0.022 | 7.102 ± 0.031 |
5.751 ± 0.038 | 10.638 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.446 ± 0.018 | 4.592 ± 0.033 |
3.646 ± 0.028 | 4.773 ± 0.039 |
3.747 ± 0.026 | 6.813 ± 0.034 |
5.597 ± 0.033 | 6.744 ± 0.036 |
1.117 ± 0.016 | 3.122 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
