
Chondrus crispus (Carrageen Irish moss) (Polymorpha crispa)
Taxonomy: cellular organisms; Eukaryota; Rhodophyta; Florideophyceae; Rhodymeniophycidae; Gigartinales; Gigartinaceae; Chondrus
Average proteome isoelectric point is 7.41
Get precalculated fractions of proteins
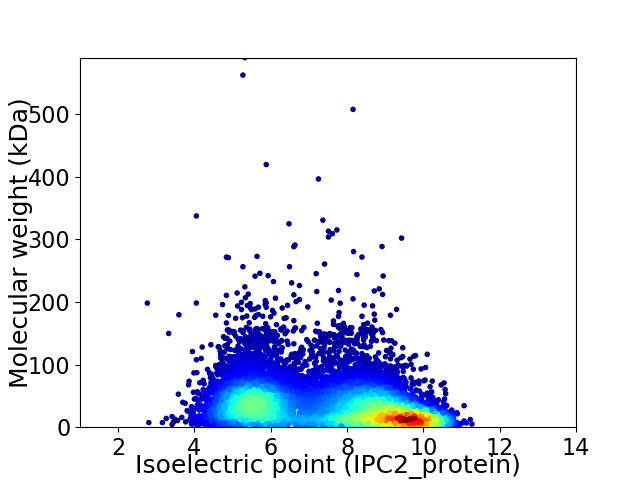
Virtual 2D-PAGE plot for 9598 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7QPH3|R7QPH3_CHOCR Symplekin-like OS=Chondrus crispus OX=2769 GN=CHC_T00010348001 PE=4 SV=1
MM1 pKa = 7.47EE2 pKa = 4.63WNEE5 pKa = 3.9LVEE8 pKa = 4.99GSDD11 pKa = 4.77LEE13 pKa = 5.28ADD15 pKa = 4.34GYY17 pKa = 6.96TTFIEE22 pKa = 4.48VQVNRR27 pKa = 11.84TLAPAVSLIAPTGPFGTQGTDD48 pKa = 3.13QVVVYY53 pKa = 9.55IDD55 pKa = 3.6NVAEE59 pKa = 4.57GEE61 pKa = 4.21DD62 pKa = 5.18DD63 pKa = 3.9PTSWTYY69 pKa = 11.48DD70 pKa = 3.45LSILFADD77 pKa = 4.85GAKK80 pKa = 10.12SATVDD85 pKa = 3.74TPAVTQFGNGTISVAFTLPAGSGVDD110 pKa = 4.05LPWNLTITKK119 pKa = 9.27PDD121 pKa = 3.45AQVVPVVDD129 pKa = 3.48ATNPSFLFSFVSQLSVIDD147 pKa = 3.97IQPSSGPEE155 pKa = 3.5EE156 pKa = 4.4GGITVSLIGTFPGFDD171 pKa = 3.37TEE173 pKa = 4.64AVDD176 pKa = 5.1VLFDD180 pKa = 4.13GNPINPEE187 pKa = 4.72LITVIGPEE195 pKa = 4.47NITFTLPPKK204 pKa = 10.46VSIGTEE210 pKa = 3.48FDD212 pKa = 3.74VNVTVSIGAQVTSGITFSYY231 pKa = 9.98IPSILLQSLTPASGPVEE248 pKa = 4.57GGTQVTLVGQFIDD261 pKa = 3.95FTSDD265 pKa = 2.57IDD267 pKa = 3.67GTGIYY272 pKa = 10.35FGGEE276 pKa = 4.19KK277 pKa = 9.17IDD279 pKa = 3.95SDD281 pKa = 5.7LITASNSSVIVFTTPPQSFFASEE304 pKa = 3.8SVYY307 pKa = 10.64AYY309 pKa = 10.22DD310 pKa = 3.69VSVHH314 pKa = 6.13IGNSSSNSIVFSYY327 pKa = 10.35DD328 pKa = 2.74APLQITSIAPNSGTEE343 pKa = 3.91EE344 pKa = 4.41GGTDD348 pKa = 2.92ITLGGTFQAFDD359 pKa = 3.92LSNSNVYY366 pKa = 10.11IGSTKK371 pKa = 9.54IDD373 pKa = 3.23SSTIVHH379 pKa = 6.75NSTSILFITPPRR391 pKa = 11.84EE392 pKa = 3.97EE393 pKa = 4.02VGNGYY398 pKa = 7.91TYY400 pKa = 10.52NIWITIEE407 pKa = 4.25TTSSNGTSTTITSNTVTFTYY427 pKa = 10.47EE428 pKa = 3.96DD429 pKa = 3.26VGSQVSIDD437 pKa = 3.49ASGGSFNSDD446 pKa = 2.04GHH448 pKa = 6.85YY449 pKa = 10.89EE450 pKa = 4.08LGQCVEE456 pKa = 4.51SLYY459 pKa = 10.35QASISRR465 pKa = 11.84GARR468 pKa = 11.84LQNASYY474 pKa = 10.35RR475 pKa = 11.84WSLVANDD482 pKa = 4.3TEE484 pKa = 4.4TDD486 pKa = 3.13ILSAKK491 pKa = 10.46GIVTNSDD498 pKa = 3.58VLLLPYY504 pKa = 10.0TVFSEE509 pKa = 4.24EE510 pKa = 3.62NAAYY514 pKa = 9.0TLTLTVDD521 pKa = 3.12TDD523 pKa = 4.12FFSFTKK529 pKa = 10.36TLVLTQLSSQNIGVRR544 pKa = 11.84IKK546 pKa = 10.57NPRR549 pKa = 11.84PRR551 pKa = 11.84SISGPNTTLTIPADD565 pKa = 4.15LYY567 pKa = 11.67LPGCQGNVIIVNSTEE582 pKa = 3.81MTYY585 pKa = 10.11EE586 pKa = 3.67WVFRR590 pKa = 11.84GEE592 pKa = 4.48TYY594 pKa = 10.9VFSHH598 pKa = 5.93QNEE601 pKa = 4.5SAPEE605 pKa = 3.94EE606 pKa = 4.18QASPTLLGRR615 pKa = 11.84EE616 pKa = 3.78FHH618 pKa = 6.74IPQAVMEE625 pKa = 4.38YY626 pKa = 10.25GSFGISLTAYY636 pKa = 8.23FTDD639 pKa = 3.49QPSIKK644 pKa = 10.63GIDD647 pKa = 3.4TSTVVINPAALISQINGGEE666 pKa = 3.83ASQLISEE673 pKa = 4.45TGKK676 pKa = 10.13FHH678 pKa = 6.94SLWFTIPRR686 pKa = 11.84SS687 pKa = 3.52
MM1 pKa = 7.47EE2 pKa = 4.63WNEE5 pKa = 3.9LVEE8 pKa = 4.99GSDD11 pKa = 4.77LEE13 pKa = 5.28ADD15 pKa = 4.34GYY17 pKa = 6.96TTFIEE22 pKa = 4.48VQVNRR27 pKa = 11.84TLAPAVSLIAPTGPFGTQGTDD48 pKa = 3.13QVVVYY53 pKa = 9.55IDD55 pKa = 3.6NVAEE59 pKa = 4.57GEE61 pKa = 4.21DD62 pKa = 5.18DD63 pKa = 3.9PTSWTYY69 pKa = 11.48DD70 pKa = 3.45LSILFADD77 pKa = 4.85GAKK80 pKa = 10.12SATVDD85 pKa = 3.74TPAVTQFGNGTISVAFTLPAGSGVDD110 pKa = 4.05LPWNLTITKK119 pKa = 9.27PDD121 pKa = 3.45AQVVPVVDD129 pKa = 3.48ATNPSFLFSFVSQLSVIDD147 pKa = 3.97IQPSSGPEE155 pKa = 3.5EE156 pKa = 4.4GGITVSLIGTFPGFDD171 pKa = 3.37TEE173 pKa = 4.64AVDD176 pKa = 5.1VLFDD180 pKa = 4.13GNPINPEE187 pKa = 4.72LITVIGPEE195 pKa = 4.47NITFTLPPKK204 pKa = 10.46VSIGTEE210 pKa = 3.48FDD212 pKa = 3.74VNVTVSIGAQVTSGITFSYY231 pKa = 9.98IPSILLQSLTPASGPVEE248 pKa = 4.57GGTQVTLVGQFIDD261 pKa = 3.95FTSDD265 pKa = 2.57IDD267 pKa = 3.67GTGIYY272 pKa = 10.35FGGEE276 pKa = 4.19KK277 pKa = 9.17IDD279 pKa = 3.95SDD281 pKa = 5.7LITASNSSVIVFTTPPQSFFASEE304 pKa = 3.8SVYY307 pKa = 10.64AYY309 pKa = 10.22DD310 pKa = 3.69VSVHH314 pKa = 6.13IGNSSSNSIVFSYY327 pKa = 10.35DD328 pKa = 2.74APLQITSIAPNSGTEE343 pKa = 3.91EE344 pKa = 4.41GGTDD348 pKa = 2.92ITLGGTFQAFDD359 pKa = 3.92LSNSNVYY366 pKa = 10.11IGSTKK371 pKa = 9.54IDD373 pKa = 3.23SSTIVHH379 pKa = 6.75NSTSILFITPPRR391 pKa = 11.84EE392 pKa = 3.97EE393 pKa = 4.02VGNGYY398 pKa = 7.91TYY400 pKa = 10.52NIWITIEE407 pKa = 4.25TTSSNGTSTTITSNTVTFTYY427 pKa = 10.47EE428 pKa = 3.96DD429 pKa = 3.26VGSQVSIDD437 pKa = 3.49ASGGSFNSDD446 pKa = 2.04GHH448 pKa = 6.85YY449 pKa = 10.89EE450 pKa = 4.08LGQCVEE456 pKa = 4.51SLYY459 pKa = 10.35QASISRR465 pKa = 11.84GARR468 pKa = 11.84LQNASYY474 pKa = 10.35RR475 pKa = 11.84WSLVANDD482 pKa = 4.3TEE484 pKa = 4.4TDD486 pKa = 3.13ILSAKK491 pKa = 10.46GIVTNSDD498 pKa = 3.58VLLLPYY504 pKa = 10.0TVFSEE509 pKa = 4.24EE510 pKa = 3.62NAAYY514 pKa = 9.0TLTLTVDD521 pKa = 3.12TDD523 pKa = 4.12FFSFTKK529 pKa = 10.36TLVLTQLSSQNIGVRR544 pKa = 11.84IKK546 pKa = 10.57NPRR549 pKa = 11.84PRR551 pKa = 11.84SISGPNTTLTIPADD565 pKa = 4.15LYY567 pKa = 11.67LPGCQGNVIIVNSTEE582 pKa = 3.81MTYY585 pKa = 10.11EE586 pKa = 3.67WVFRR590 pKa = 11.84GEE592 pKa = 4.48TYY594 pKa = 10.9VFSHH598 pKa = 5.93QNEE601 pKa = 4.5SAPEE605 pKa = 3.94EE606 pKa = 4.18QASPTLLGRR615 pKa = 11.84EE616 pKa = 3.78FHH618 pKa = 6.74IPQAVMEE625 pKa = 4.38YY626 pKa = 10.25GSFGISLTAYY636 pKa = 8.23FTDD639 pKa = 3.49QPSIKK644 pKa = 10.63GIDD647 pKa = 3.4TSTVVINPAALISQINGGEE666 pKa = 3.83ASQLISEE673 pKa = 4.45TGKK676 pKa = 10.13FHH678 pKa = 6.94SLWFTIPRR686 pKa = 11.84SS687 pKa = 3.52
Molecular weight: 73.25 kDa
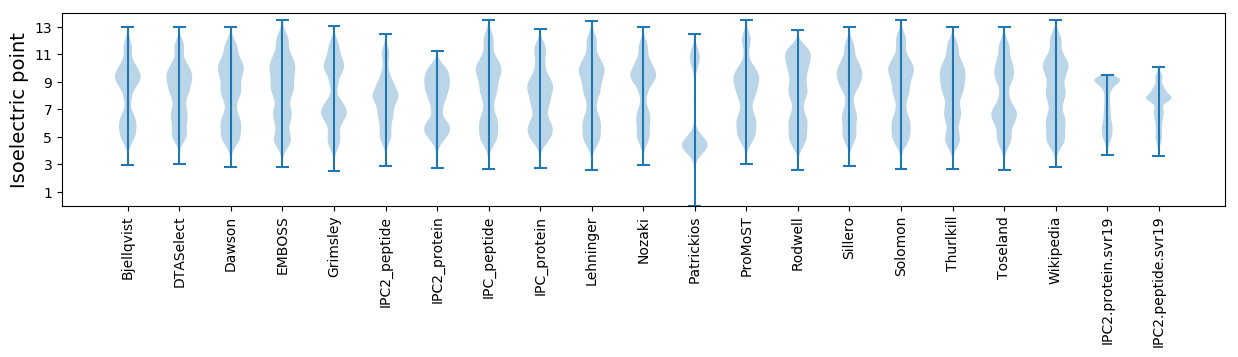
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7Q251|R7Q251_CHOCR Uncharacterized protein OS=Chondrus crispus OX=2769 GN=CHC_T00001368001 PE=4 SV=1
MM1 pKa = 7.47TKK3 pKa = 9.12GTNRR7 pKa = 11.84KK8 pKa = 9.3KK9 pKa = 9.82IRR11 pKa = 11.84KK12 pKa = 8.2AGFRR16 pKa = 11.84ARR18 pKa = 11.84MKK20 pKa = 8.98TVSGRR25 pKa = 11.84RR26 pKa = 11.84IIKK29 pKa = 9.51ARR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 8.44KK35 pKa = 10.03RR36 pKa = 11.84KK37 pKa = 9.82RR38 pKa = 11.84IGLL41 pKa = 3.8
MM1 pKa = 7.47TKK3 pKa = 9.12GTNRR7 pKa = 11.84KK8 pKa = 9.3KK9 pKa = 9.82IRR11 pKa = 11.84KK12 pKa = 8.2AGFRR16 pKa = 11.84ARR18 pKa = 11.84MKK20 pKa = 8.98TVSGRR25 pKa = 11.84RR26 pKa = 11.84IIKK29 pKa = 9.51ARR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 8.44KK35 pKa = 10.03RR36 pKa = 11.84KK37 pKa = 9.82RR38 pKa = 11.84IGLL41 pKa = 3.8
Molecular weight: 4.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3411166 |
10 |
5324 |
355.4 |
39.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.083 ± 0.027 | 1.749 ± 0.015 |
5.399 ± 0.022 | 6.123 ± 0.033 |
3.618 ± 0.019 | 6.788 ± 0.028 |
2.531 ± 0.015 | 4.332 ± 0.02 |
4.763 ± 0.024 | 8.995 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.328 ± 0.011 | 3.295 ± 0.017 |
5.519 ± 0.04 | 3.446 ± 0.02 |
7.486 ± 0.026 | 8.141 ± 0.034 |
5.424 ± 0.019 | 7.021 ± 0.022 |
1.171 ± 0.009 | 2.315 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
