
Torque teno virus 10
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 9.21
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
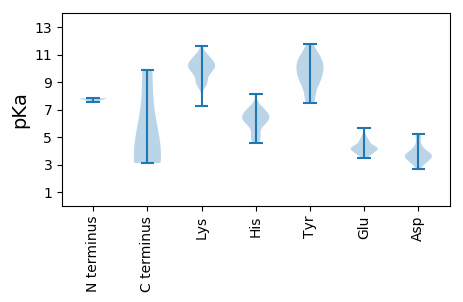
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|E3TH04|E3TH04_9VIRU Capsid protein OS=Torque teno virus 10 OX=687349 PE=3 SV=1
MM1 pKa = 7.81AEE3 pKa = 3.58FMLPVRR9 pKa = 11.84RR10 pKa = 11.84EE11 pKa = 3.67EE12 pKa = 3.79PRR14 pKa = 11.84RR15 pKa = 11.84GIRR18 pKa = 11.84TSRR21 pKa = 11.84GRR23 pKa = 11.84VPEE26 pKa = 4.17VSLHH30 pKa = 5.18TAVKK34 pKa = 9.84GQFGLGTGRR43 pKa = 11.84AMGKK47 pKa = 9.74ALKK50 pKa = 10.28KK51 pKa = 11.02AMFLGKK57 pKa = 9.37LHH59 pKa = 6.44RR60 pKa = 11.84KK61 pKa = 8.87KK62 pKa = 10.46RR63 pKa = 11.84ALSLHH68 pKa = 6.22GLPATKK74 pKa = 10.08KK75 pKa = 10.44KK76 pKa = 10.28PPPDD80 pKa = 3.37MNYY83 pKa = 9.21WRR85 pKa = 11.84PPVHH89 pKa = 6.27NVPGLEE95 pKa = 3.98RR96 pKa = 11.84LWYY99 pKa = 8.59EE100 pKa = 4.09SVHH103 pKa = 6.23RR104 pKa = 11.84SHH106 pKa = 7.16AAVCGCGDD114 pKa = 4.23FVRR117 pKa = 11.84HH118 pKa = 4.6ITALAEE124 pKa = 4.44RR125 pKa = 11.84YY126 pKa = 7.86GHH128 pKa = 6.91PGGPRR133 pKa = 11.84APGAPGIGGNPNSPPIRR150 pKa = 11.84RR151 pKa = 11.84ARR153 pKa = 11.84HH154 pKa = 4.98PAAAPEE160 pKa = 4.31PPAGNQPPALPWHH173 pKa = 6.7GDD175 pKa = 3.08GGNEE179 pKa = 4.13GASGGGDD186 pKa = 3.51DD187 pKa = 5.03AGLVADD193 pKa = 5.23FANDD197 pKa = 3.69GLDD200 pKa = 3.61EE201 pKa = 4.46LVAALDD207 pKa = 3.78EE208 pKa = 4.67EE209 pKa = 4.8EE210 pKa = 4.1
MM1 pKa = 7.81AEE3 pKa = 3.58FMLPVRR9 pKa = 11.84RR10 pKa = 11.84EE11 pKa = 3.67EE12 pKa = 3.79PRR14 pKa = 11.84RR15 pKa = 11.84GIRR18 pKa = 11.84TSRR21 pKa = 11.84GRR23 pKa = 11.84VPEE26 pKa = 4.17VSLHH30 pKa = 5.18TAVKK34 pKa = 9.84GQFGLGTGRR43 pKa = 11.84AMGKK47 pKa = 9.74ALKK50 pKa = 10.28KK51 pKa = 11.02AMFLGKK57 pKa = 9.37LHH59 pKa = 6.44RR60 pKa = 11.84KK61 pKa = 8.87KK62 pKa = 10.46RR63 pKa = 11.84ALSLHH68 pKa = 6.22GLPATKK74 pKa = 10.08KK75 pKa = 10.44KK76 pKa = 10.28PPPDD80 pKa = 3.37MNYY83 pKa = 9.21WRR85 pKa = 11.84PPVHH89 pKa = 6.27NVPGLEE95 pKa = 3.98RR96 pKa = 11.84LWYY99 pKa = 8.59EE100 pKa = 4.09SVHH103 pKa = 6.23RR104 pKa = 11.84SHH106 pKa = 7.16AAVCGCGDD114 pKa = 4.23FVRR117 pKa = 11.84HH118 pKa = 4.6ITALAEE124 pKa = 4.44RR125 pKa = 11.84YY126 pKa = 7.86GHH128 pKa = 6.91PGGPRR133 pKa = 11.84APGAPGIGGNPNSPPIRR150 pKa = 11.84RR151 pKa = 11.84ARR153 pKa = 11.84HH154 pKa = 4.98PAAAPEE160 pKa = 4.31PPAGNQPPALPWHH173 pKa = 6.7GDD175 pKa = 3.08GGNEE179 pKa = 4.13GASGGGDD186 pKa = 3.51DD187 pKa = 5.03AGLVADD193 pKa = 5.23FANDD197 pKa = 3.69GLDD200 pKa = 3.61EE201 pKa = 4.46LVAALDD207 pKa = 3.78EE208 pKa = 4.67EE209 pKa = 4.8EE210 pKa = 4.1
Molecular weight: 22.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E3TH02|E3TH02_9VIRU ORF2/3 OS=Torque teno virus 10 OX=687349 PE=4 SV=1
MM1 pKa = 7.81AEE3 pKa = 3.58FMLPVRR9 pKa = 11.84RR10 pKa = 11.84EE11 pKa = 3.67EE12 pKa = 3.79PRR14 pKa = 11.84RR15 pKa = 11.84GIRR18 pKa = 11.84TSRR21 pKa = 11.84GRR23 pKa = 11.84VPEE26 pKa = 4.17VSLHH30 pKa = 5.18TAVKK34 pKa = 9.84GQFGLGTGRR43 pKa = 11.84AMGKK47 pKa = 9.74ALKK50 pKa = 10.28KK51 pKa = 11.02AMFLGKK57 pKa = 9.37LHH59 pKa = 6.44RR60 pKa = 11.84KK61 pKa = 8.87KK62 pKa = 10.46RR63 pKa = 11.84ALSLHH68 pKa = 6.22GLPATKK74 pKa = 10.08KK75 pKa = 10.44KK76 pKa = 10.28PPPDD80 pKa = 3.37MNYY83 pKa = 9.21WRR85 pKa = 11.84PPVHH89 pKa = 6.27NVPGLEE95 pKa = 3.98RR96 pKa = 11.84LWYY99 pKa = 8.59EE100 pKa = 4.09SVHH103 pKa = 6.23RR104 pKa = 11.84SHH106 pKa = 7.16AAVCGCGDD114 pKa = 4.23FVRR117 pKa = 11.84HH118 pKa = 4.6ITALAEE124 pKa = 4.44RR125 pKa = 11.84YY126 pKa = 7.86GHH128 pKa = 6.91PGGPRR133 pKa = 11.84APGAPGIGGNPNSPPIRR150 pKa = 11.84RR151 pKa = 11.84ARR153 pKa = 11.84HH154 pKa = 4.98PAAAPEE160 pKa = 4.31PPAGNQPPALPWHH173 pKa = 6.7GDD175 pKa = 3.08GGNEE179 pKa = 4.13GASGGGDD186 pKa = 3.51DD187 pKa = 5.03AGLVADD193 pKa = 5.23FANDD197 pKa = 3.69GLDD200 pKa = 3.61EE201 pKa = 4.46LVAALDD207 pKa = 3.81EE208 pKa = 4.68EE209 pKa = 5.03EE210 pKa = 4.29SQKK213 pKa = 8.89TQGRR217 pKa = 11.84PRR219 pKa = 11.84ANPTARR225 pKa = 11.84KK226 pKa = 9.38ALRR229 pKa = 11.84FTPKK233 pKa = 10.06RR234 pKa = 11.84IEE236 pKa = 3.88AVGDD240 pKa = 3.12QRR242 pKa = 11.84RR243 pKa = 11.84RR244 pKa = 11.84EE245 pKa = 4.06RR246 pKa = 11.84SRR248 pKa = 11.84SPARR252 pKa = 11.84RR253 pKa = 11.84DD254 pKa = 3.22GGGAPQTATPPQPQRR269 pKa = 11.84AAATPKK275 pKa = 10.2GPPVRR280 pKa = 11.84LPAANKK286 pKa = 9.11DD287 pKa = 3.58AAGGSHH293 pKa = 6.63RR294 pKa = 11.84PIPTVGPSQWLFPEE308 pKa = 5.24RR309 pKa = 11.84KK310 pKa = 8.94PKK312 pKa = 10.17PPPSAGDD319 pKa = 3.03WAMEE323 pKa = 3.8YY324 pKa = 10.2LACKK328 pKa = 9.9IFNRR332 pKa = 11.84PPRR335 pKa = 11.84THH337 pKa = 6.48LTDD340 pKa = 3.8PPFYY344 pKa = 9.94PYY346 pKa = 10.68CKK348 pKa = 9.98NNYY351 pKa = 9.41NVTFQLNYY359 pKa = 10.06KK360 pKa = 9.85
MM1 pKa = 7.81AEE3 pKa = 3.58FMLPVRR9 pKa = 11.84RR10 pKa = 11.84EE11 pKa = 3.67EE12 pKa = 3.79PRR14 pKa = 11.84RR15 pKa = 11.84GIRR18 pKa = 11.84TSRR21 pKa = 11.84GRR23 pKa = 11.84VPEE26 pKa = 4.17VSLHH30 pKa = 5.18TAVKK34 pKa = 9.84GQFGLGTGRR43 pKa = 11.84AMGKK47 pKa = 9.74ALKK50 pKa = 10.28KK51 pKa = 11.02AMFLGKK57 pKa = 9.37LHH59 pKa = 6.44RR60 pKa = 11.84KK61 pKa = 8.87KK62 pKa = 10.46RR63 pKa = 11.84ALSLHH68 pKa = 6.22GLPATKK74 pKa = 10.08KK75 pKa = 10.44KK76 pKa = 10.28PPPDD80 pKa = 3.37MNYY83 pKa = 9.21WRR85 pKa = 11.84PPVHH89 pKa = 6.27NVPGLEE95 pKa = 3.98RR96 pKa = 11.84LWYY99 pKa = 8.59EE100 pKa = 4.09SVHH103 pKa = 6.23RR104 pKa = 11.84SHH106 pKa = 7.16AAVCGCGDD114 pKa = 4.23FVRR117 pKa = 11.84HH118 pKa = 4.6ITALAEE124 pKa = 4.44RR125 pKa = 11.84YY126 pKa = 7.86GHH128 pKa = 6.91PGGPRR133 pKa = 11.84APGAPGIGGNPNSPPIRR150 pKa = 11.84RR151 pKa = 11.84ARR153 pKa = 11.84HH154 pKa = 4.98PAAAPEE160 pKa = 4.31PPAGNQPPALPWHH173 pKa = 6.7GDD175 pKa = 3.08GGNEE179 pKa = 4.13GASGGGDD186 pKa = 3.51DD187 pKa = 5.03AGLVADD193 pKa = 5.23FANDD197 pKa = 3.69GLDD200 pKa = 3.61EE201 pKa = 4.46LVAALDD207 pKa = 3.81EE208 pKa = 4.68EE209 pKa = 5.03EE210 pKa = 4.29SQKK213 pKa = 8.89TQGRR217 pKa = 11.84PRR219 pKa = 11.84ANPTARR225 pKa = 11.84KK226 pKa = 9.38ALRR229 pKa = 11.84FTPKK233 pKa = 10.06RR234 pKa = 11.84IEE236 pKa = 3.88AVGDD240 pKa = 3.12QRR242 pKa = 11.84RR243 pKa = 11.84RR244 pKa = 11.84EE245 pKa = 4.06RR246 pKa = 11.84SRR248 pKa = 11.84SPARR252 pKa = 11.84RR253 pKa = 11.84DD254 pKa = 3.22GGGAPQTATPPQPQRR269 pKa = 11.84AAATPKK275 pKa = 10.2GPPVRR280 pKa = 11.84LPAANKK286 pKa = 9.11DD287 pKa = 3.58AAGGSHH293 pKa = 6.63RR294 pKa = 11.84PIPTVGPSQWLFPEE308 pKa = 5.24RR309 pKa = 11.84KK310 pKa = 8.94PKK312 pKa = 10.17PPPSAGDD319 pKa = 3.03WAMEE323 pKa = 3.8YY324 pKa = 10.2LACKK328 pKa = 9.9IFNRR332 pKa = 11.84PPRR335 pKa = 11.84THH337 pKa = 6.48LTDD340 pKa = 3.8PPFYY344 pKa = 9.94PYY346 pKa = 10.68CKK348 pKa = 9.98NNYY351 pKa = 9.41NVTFQLNYY359 pKa = 10.06KK360 pKa = 9.85
Molecular weight: 39.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1665 |
210 |
744 |
416.3 |
46.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.808 ± 2.01 | 1.381 ± 0.14 |
4.024 ± 0.301 | 5.045 ± 0.37 |
3.363 ± 0.787 | 8.589 ± 1.843 |
3.183 ± 0.481 | 3.063 ± 0.839 |
6.306 ± 0.486 | 7.568 ± 0.419 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.802 ± 0.149 | 4.565 ± 0.358 |
9.369 ± 1.707 | 3.604 ± 1.106 |
9.489 ± 0.224 | 6.066 ± 1.454 |
5.045 ± 0.834 | 4.745 ± 0.199 |
2.102 ± 0.641 | 2.883 ± 0.903 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
