
Aspergillus versicolor CBS 583.65
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Aspergillaceae; Aspergillus; Aspergillus subgen. Nidulantes; Aspergillus versicolor
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
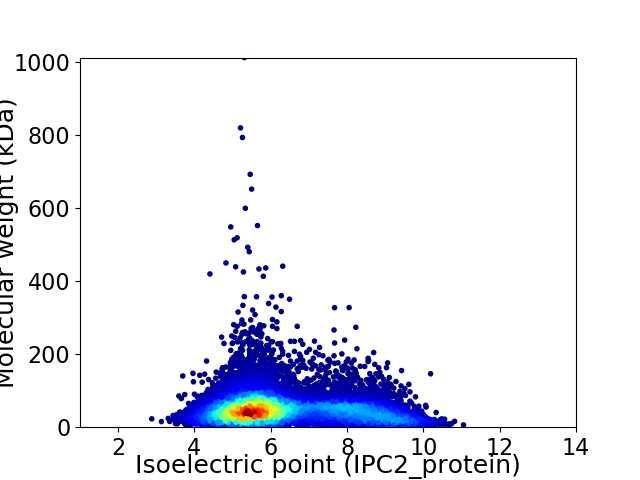
Virtual 2D-PAGE plot for 13222 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L9PMJ3|A0A1L9PMJ3_ASPVE Uncharacterized protein OS=Aspergillus versicolor CBS 583.65 OX=1036611 GN=ASPVEDRAFT_131628 PE=4 SV=1
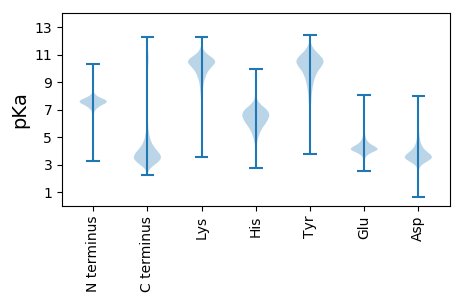
MM1 pKa = 6.74THH3 pKa = 5.71STLGSLYY10 pKa = 10.74LFLAPILSVLAQDD23 pKa = 5.33LPADD27 pKa = 3.7LTGCTEE33 pKa = 3.94VDD35 pKa = 4.12CPNDD39 pKa = 3.23GWDD42 pKa = 3.43SCTVADD48 pKa = 4.2EE49 pKa = 4.62TYY51 pKa = 11.14AGIGLSGIDD60 pKa = 4.17SVPDD64 pKa = 3.51SLDD67 pKa = 3.88GISLVKK73 pKa = 10.25GVHH76 pKa = 5.8VEE78 pKa = 4.03DD79 pKa = 5.71DD80 pKa = 3.81EE81 pKa = 6.23DD82 pKa = 6.29RR83 pKa = 11.84GDD85 pKa = 3.77TDD87 pKa = 3.37NGADD91 pKa = 3.16KK92 pKa = 10.38TRR94 pKa = 11.84PFRR97 pKa = 11.84SVYY100 pKa = 10.84YY101 pKa = 10.41LGTPANVNVDD111 pKa = 4.19DD112 pKa = 5.19LSGCAVVFNDD122 pKa = 4.34PPTGHH127 pKa = 6.46FNKK130 pKa = 10.16PLINDD135 pKa = 3.67SVNVNTRR142 pKa = 11.84AAYY145 pKa = 7.08GTCPDD150 pKa = 4.94VIQPDD155 pKa = 4.7CIDD158 pKa = 3.95KK159 pKa = 9.36LTEE162 pKa = 3.68QARR165 pKa = 11.84KK166 pKa = 7.28TKK168 pKa = 10.78YY169 pKa = 10.09NGSSPCSTLQSALQDD184 pKa = 3.81NPPDD188 pKa = 3.68EE189 pKa = 5.71CSDD192 pKa = 3.57LTGGGDD198 pKa = 3.67GLGKK202 pKa = 10.21FDD204 pKa = 5.61VISLGNLSTISKK216 pKa = 10.05SDD218 pKa = 3.64NASSDD223 pKa = 3.6CWPVLPKK230 pKa = 10.73SNNLAQLTDD239 pKa = 3.61DD240 pKa = 3.85TAMGNYY246 pKa = 8.5TNEE249 pKa = 3.84GLLAEE254 pKa = 4.75MYY256 pKa = 10.39KK257 pKa = 8.78ITPVLTVFTSGNNSNSLVNEE277 pKa = 4.5TSASLTCLKK286 pKa = 10.78VVTTPNIDD294 pKa = 3.27NGTDD298 pKa = 3.28ADD300 pKa = 4.11TDD302 pKa = 3.77TDD304 pKa = 3.46TGAAALSPANMVAVTLGALATAAFALLL331 pKa = 3.78
MM1 pKa = 6.74THH3 pKa = 5.71STLGSLYY10 pKa = 10.74LFLAPILSVLAQDD23 pKa = 5.33LPADD27 pKa = 3.7LTGCTEE33 pKa = 3.94VDD35 pKa = 4.12CPNDD39 pKa = 3.23GWDD42 pKa = 3.43SCTVADD48 pKa = 4.2EE49 pKa = 4.62TYY51 pKa = 11.14AGIGLSGIDD60 pKa = 4.17SVPDD64 pKa = 3.51SLDD67 pKa = 3.88GISLVKK73 pKa = 10.25GVHH76 pKa = 5.8VEE78 pKa = 4.03DD79 pKa = 5.71DD80 pKa = 3.81EE81 pKa = 6.23DD82 pKa = 6.29RR83 pKa = 11.84GDD85 pKa = 3.77TDD87 pKa = 3.37NGADD91 pKa = 3.16KK92 pKa = 10.38TRR94 pKa = 11.84PFRR97 pKa = 11.84SVYY100 pKa = 10.84YY101 pKa = 10.41LGTPANVNVDD111 pKa = 4.19DD112 pKa = 5.19LSGCAVVFNDD122 pKa = 4.34PPTGHH127 pKa = 6.46FNKK130 pKa = 10.16PLINDD135 pKa = 3.67SVNVNTRR142 pKa = 11.84AAYY145 pKa = 7.08GTCPDD150 pKa = 4.94VIQPDD155 pKa = 4.7CIDD158 pKa = 3.95KK159 pKa = 9.36LTEE162 pKa = 3.68QARR165 pKa = 11.84KK166 pKa = 7.28TKK168 pKa = 10.78YY169 pKa = 10.09NGSSPCSTLQSALQDD184 pKa = 3.81NPPDD188 pKa = 3.68EE189 pKa = 5.71CSDD192 pKa = 3.57LTGGGDD198 pKa = 3.67GLGKK202 pKa = 10.21FDD204 pKa = 5.61VISLGNLSTISKK216 pKa = 10.05SDD218 pKa = 3.64NASSDD223 pKa = 3.6CWPVLPKK230 pKa = 10.73SNNLAQLTDD239 pKa = 3.61DD240 pKa = 3.85TAMGNYY246 pKa = 8.5TNEE249 pKa = 3.84GLLAEE254 pKa = 4.75MYY256 pKa = 10.39KK257 pKa = 8.78ITPVLTVFTSGNNSNSLVNEE277 pKa = 4.5TSASLTCLKK286 pKa = 10.78VVTTPNIDD294 pKa = 3.27NGTDD298 pKa = 3.28ADD300 pKa = 4.11TDD302 pKa = 3.77TDD304 pKa = 3.46TGAAALSPANMVAVTLGALATAAFALLL331 pKa = 3.78
Molecular weight: 34.52 kDa
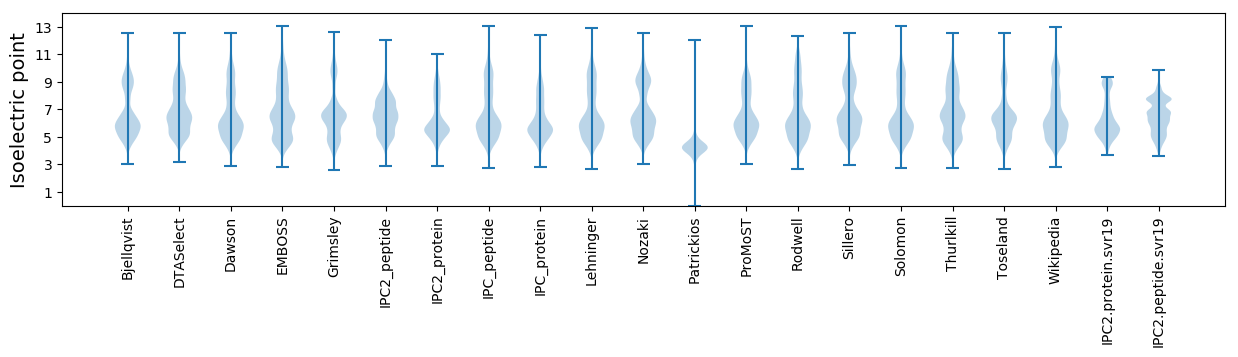
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L9PC49|A0A1L9PC49_ASPVE Uncharacterized protein OS=Aspergillus versicolor CBS 583.65 OX=1036611 GN=ASPVEDRAFT_25939 PE=4 SV=1
MM1 pKa = 7.94PSQKK5 pKa = 10.41SFRR8 pKa = 11.84TKK10 pKa = 10.38QKK12 pKa = 9.84LAKK15 pKa = 9.55AQKK18 pKa = 8.59QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.89RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.14WRR45 pKa = 11.84KK46 pKa = 7.41TRR48 pKa = 11.84LGII51 pKa = 4.46
MM1 pKa = 7.94PSQKK5 pKa = 10.41SFRR8 pKa = 11.84TKK10 pKa = 10.38QKK12 pKa = 9.84LAKK15 pKa = 9.55AQKK18 pKa = 8.59QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.89RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.14WRR45 pKa = 11.84KK46 pKa = 7.41TRR48 pKa = 11.84LGII51 pKa = 4.46
Molecular weight: 6.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6349617 |
49 |
9209 |
480.2 |
53.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.448 ± 0.017 | 1.341 ± 0.007 |
5.635 ± 0.015 | 5.999 ± 0.02 |
3.861 ± 0.012 | 7.028 ± 0.018 |
2.396 ± 0.009 | 5.082 ± 0.013 |
4.399 ± 0.015 | 9.147 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.112 ± 0.009 | 3.661 ± 0.01 |
5.982 ± 0.022 | 3.965 ± 0.015 |
6.042 ± 0.018 | 8.256 ± 0.022 |
5.918 ± 0.014 | 6.273 ± 0.014 |
1.542 ± 0.008 | 2.913 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
