
Desulforhopalus singaporensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobulbaceae; Desulforhopalus
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
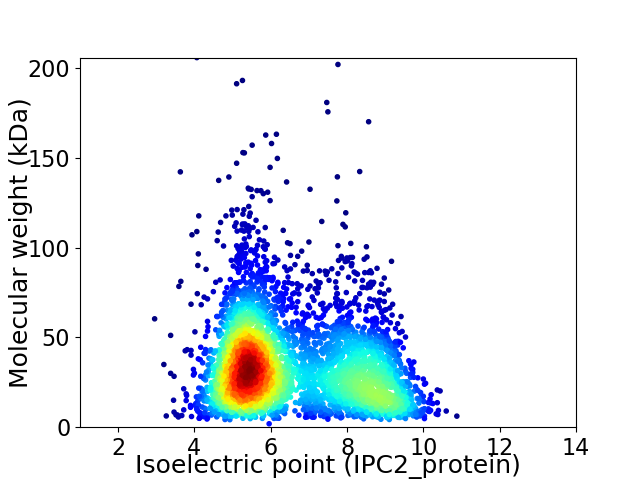
Virtual 2D-PAGE plot for 4348 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H0R7U2|A0A1H0R7U2_9DELT Protein HflK OS=Desulforhopalus singaporensis OX=91360 GN=SAMN05660330_02210 PE=3 SV=1
DDD2 pKa = 3.94EE3 pKa = 4.43TTRR6 pKa = 11.84GEEE9 pKa = 3.56DDD11 pKa = 3.64YY12 pKa = 10.18YYY14 pKa = 11.14KK15 pKa = 10.91QIVDDD20 pKa = 3.19KK21 pKa = 10.37GNVGNVDDD29 pKa = 3.79QDDD32 pKa = 3.28TIDDD36 pKa = 3.61SGPQDDD42 pKa = 3.46TIDDD46 pKa = 3.28TGITEEE52 pKa = 4.07DD53 pKa = 3.74GLVADDD59 pKa = 5.62DD60 pKa = 5.19YY61 pKa = 9.79YY62 pKa = 11.4NDDD65 pKa = 3.17TLVVSGVLGVALLAGEEE82 pKa = 4.41VQVQLDDD89 pKa = 3.25GNWYYY94 pKa = 7.66ATIDDD99 pKa = 3.54TGTGWSFDDD108 pKa = 3.92SSTTLDDD115 pKa = 3.43EE116 pKa = 4.77DDD118 pKa = 3.84HH119 pKa = 5.85FNAKKK124 pKa = 9.09VNAAGNDDD132 pKa = 3.6ATTSKKK138 pKa = 9.85VTVDDD143 pKa = 3.32TADDD147 pKa = 3.51AVAEEE152 pKa = 4.41TSIADDD158 pKa = 3.37DD159 pKa = 3.65GMVTDDD165 pKa = 5.62DD166 pKa = 4.25VTNDDD171 pKa = 3.27TLTVYYY177 pKa = 8.7TLDDD181 pKa = 3.12KK182 pKa = 11.31LGEEE186 pKa = 4.03SYYY189 pKa = 10.57EEE191 pKa = 3.6WGARR195 pKa = 11.84EEE197 pKa = 3.54QTPVKKK203 pKa = 10.18GVATVDDD210 pKa = 3.42TGTGWSFDDD219 pKa = 3.59SGNPHHH225 pKa = 6.77EE226 pKa = 5.41DD227 pKa = 3.14EEE229 pKa = 4.5YY230 pKa = 9.33YYY232 pKa = 10.1YYY234 pKa = 8.5EE235 pKa = 5.6RR236 pKa = 11.84VFDDD240 pKa = 3.49GGNSGSVDDD249 pKa = 3.52KKK251 pKa = 11.22VQIDDD256 pKa = 3.64VGPEEE261 pKa = 4.09LNLDDD266 pKa = 3.3TFEEE270 pKa = 4.34LTDDD274 pKa = 4.02YYY276 pKa = 11.0HH277 pKa = 7.68LDDD280 pKa = 5.06DDD282 pKa = 4.43EEE284 pKa = 4.71DD285 pKa = 4.45VSGINNDDD293 pKa = 4.54ITRR296 pKa = 11.84DDD298 pKa = 3.72YY299 pKa = 10.76ITIAGEEE306 pKa = 4.04SSSLQAGEEE315 pKa = 4.26YY316 pKa = 10.09QISTDDD322 pKa = 3.58EEE324 pKa = 3.98WEEE327 pKa = 4.67LLQDDD332 pKa = 4.79GKKK335 pKa = 9.08WSWKKK340 pKa = 10.11VDDD343 pKa = 3.16DDD345 pKa = 4.07YY346 pKa = 11.56VDDD349 pKa = 3.55TTTYYY354 pKa = 11.4YY355 pKa = 10.92RR356 pKa = 11.84LVDDD360 pKa = 3.85AGNEEE365 pKa = 4.04TGTVFKKK372 pKa = 10.29DDD374 pKa = 2.54GYYY377 pKa = 10.17EE378 pKa = 4.12TVDDD382 pKa = 3.62TAPDDD387 pKa = 5.56ILVAPGGVPEEE398 pKa = 4.34TSTTEEE404 pKa = 3.49EE405 pKa = 4.06TFDDD409 pKa = 3.13NTYYY413 pKa = 10.17KKK415 pKa = 10.19EEE417 pKa = 4.21NCYYY421 pKa = 8.1ALYYY425 pKa = 10.74DDD427 pKa = 3.59ANDDD431 pKa = 3.56SYYY434 pKa = 10.93LGVDDD439 pKa = 3.62AIHHH443 pKa = 5.38YY444 pKa = 7.9QADDD448 pKa = 3.38SGNWSMTTKKK458 pKa = 10.81PAGSNNLTFAVWDDD472 pKa = 3.68EE473 pKa = 4.17GNRR476 pKa = 11.84SQFGPSTSVGVSEEE490 pKa = 4.58TGSEEE495 pKa = 4.7DDD497 pKa = 3.45LTWGGTVNPGLGLNSAAVTINEEE520 pKa = 4.32DD521 pKa = 3.76LWSFFQSSAALSGTTTANAGWVFAAKKK548 pKa = 10.44QDDD551 pKa = 3.76DD552 pKa = 4.06YY553 pKa = 11.98ATYYY557 pKa = 10.77EEE559 pKa = 4.22PRR561 pKa = 11.84DDD563 pKa = 3.82TSAAYYY569 pKa = 9.85EE570 pKa = 4.11FVNAALFADDD580 pKa = 4.24YY581 pKa = 11.14RR582 pKa = 11.84DDD584 pKa = 2.95YYY586 pKa = 11.63DDD588 pKa = 4.08ISQVSSYYY596 pKa = 11.46NASRR600 pKa = 11.84TAMWTNNGDDD610 pKa = 3.43TYYY613 pKa = 9.91PGSINQGTLNHHH625 pKa = 6.72GGAIAYYY632 pKa = 9.03DD633 pKa = 3.84KK634 pKa = 11.43EE635 pKa = 4.33DDD637 pKa = 3.72YYY639 pKa = 11.47DDD641 pKa = 3.96VLADDD646 pKa = 3.86EEE648 pKa = 4.4DDD650 pKa = 3.32LSFLKKK656 pKa = 10.97VDDD659 pKa = 3.81DD660 pKa = 4.75SGGRR664 pKa = 11.84KKK666 pKa = 7.17TYYY669 pKa = 8.53EE670 pKa = 4.14NAGYYY675 pKa = 10.84YY676 pKa = 9.95GVPAGVDDD684 pKa = 2.92YYY686 pKa = 11.04SILHHH691 pKa = 5.75EE692 pKa = 4.38SAVDDD697 pKa = 4.4DDD699 pKa = 3.93DDD701 pKa = 3.84SVDDD705 pKa = 3.38AAHHH709 pKa = 5.36DDD711 pKa = 3.18LGGGNPGRR719 pKa = 11.84VLTVFHHH726 pKa = 7.07EEE728 pKa = 3.94NDDD731 pKa = 3.59SSQQFTMQDDD741 pKa = 3.32YY742 pKa = 9.91NVFRR746 pKa = 11.84NDDD749 pKa = 3.21YYY751 pKa = 11.61DDD753 pKa = 3.79YY754 pKa = 11.45NLSYYY759 pKa = 11.81MTWGDDD765 pKa = 3.75YY766 pKa = 11.2DD767 pKa = 4.22DDD769 pKa = 4.22YYY771 pKa = 11.66DDD773 pKa = 4.19FLSRR777 pKa = 11.84GNIKKK782 pKa = 10.67GVGNSDDD789 pKa = 3.29EE790 pKa = 4.19RR791 pKa = 11.84IYYY794 pKa = 10.92NDDD797 pKa = 3.61SGGLKKK803 pKa = 9.12ATEEE807 pKa = 4.48DD808 pKa = 3.7TWMGDDD814 pKa = 2.87LAGGTSFAVDDD825 pKa = 3.49NHHH828 pKa = 7.16DD829 pKa = 3.31LMDDD833 pKa = 4.4VEEE836 pKa = 4.8PNSRR840 pKa = 11.84AVQFGPTLYYY850 pKa = 10.68NQGDDD855 pKa = 4.49TWEEE859 pKa = 3.81MTMTEEE865 pKa = 4.29LGSTITGGVALDDD878 pKa = 4.13YY879 pKa = 11.05WDDD882 pKa = 3.46AMDDD886 pKa = 4.42VLYYY890 pKa = 10.18PSSTSLQSGADDD902 pKa = 3.71NTAPSKKK909 pKa = 10.61VNNTNIAADDD919 pKa = 3.94TSLQLRR925 pKa = 11.84ILDDD929 pKa = 3.77EEE931 pKa = 4.32MNVYYY936 pKa = 10.3YY937 pKa = 10.98NTVKKK942 pKa = 10.82YYY944 pKa = 10.73DD945 pKa = 3.53AGNCVATTLMNPQGSGSSNAMGLVSFYYY973 pKa = 11.33LDDD976 pKa = 3.42DDD978 pKa = 3.97EE979 pKa = 5.01YYY981 pKa = 11.03VQLLRR986 pKa = 11.84TTNGEEE992 pKa = 3.91DDD994 pKa = 3.23HH995 pKa = 6.62GSVASQGGFTNSTVNSSWGGLTTGKKK1021 pKa = 9.85HHH1023 pKa = 7.78DD1024 pKa = 3.68YYY1026 pKa = 10.05LVGEEE1031 pKa = 4.77DDD1033 pKa = 3.68STASSTNEEE1042 pKa = 3.3GIVGSGYYY1050 pKa = 10.46DDD1052 pKa = 3.53FVSSAGDDD1060 pKa = 3.75DD1061 pKa = 3.32FNGSGGWSQLVISDDD1076 pKa = 4.57VWEEE1080 pKa = 4.24DDD1082 pKa = 3.56GLDDD1086 pKa = 3.22VDDD1089 pKa = 3.43YY1090 pKa = 11.22KKK1092 pKa = 10.24TGGGVQANLATGVVTGAAGNDDD1114 pKa = 3.36LINIEEE1120 pKa = 4.19LLGTSADDD1128 pKa = 3.86DD1129 pKa = 3.74FTDDD1133 pKa = 3.43AANNVFEEE1141 pKa = 4.5RR1142 pKa = 11.84GGNDDD1147 pKa = 3.05YYY1149 pKa = 11.27LTGGGRR1155 pKa = 11.84DDD1157 pKa = 3.31LLYYY1161 pKa = 10.35EE1162 pKa = 4.23LADDD1166 pKa = 4.12DDD1168 pKa = 4.05TGGNGSDDD1176 pKa = 4.24INDDD1180 pKa = 3.56TVGLVKKK1187 pKa = 10.55DD1188 pKa = 3.85ADDD1191 pKa = 4.18DDD1193 pKa = 4.22INISDDD1199 pKa = 4.06LNYYY1203 pKa = 9.84GTVGTYYY1210 pKa = 8.8DDD1212 pKa = 3.21SGTYYY1217 pKa = 8.56HH1218 pKa = 7.83DDD1220 pKa = 3.42ASEEE1224 pKa = 4.2VNDDD1228 pKa = 4.24VKKK1231 pKa = 9.88TTSGGNTEEE1240 pKa = 3.91AIDDD1244 pKa = 3.61DDD1246 pKa = 3.5AGNYYY1251 pKa = 6.54TLVTLEEE1258 pKa = 4.04TEEE1261 pKa = 4.04DDD1263 pKa = 3.96LSLLTNQQLIVEEE1276 pKa = 4.64SADDD1280 pKa = 3.51GASGAAASQQEEE1292 pKa = 4.37DD1293 pKa = 4.24EEE1295 pKa = 4.68STGSTGGDDD1304 pKa = 3.72DD1305 pKa = 3.79TASDDD1310 pKa = 4.75STTAPIADDD1319 pKa = 3.83SGNTTDDD1326 pKa = 3.24SGVVDDD1332 pKa = 3.9ADDD1335 pKa = 3.25VTTRR1339 pKa = 11.84VEEE1342 pKa = 4.34DD1343 pKa = 4.67DD1344 pKa = 5.04VTQLNSPTVI
DDD2 pKa = 3.94EE3 pKa = 4.43TTRR6 pKa = 11.84GEEE9 pKa = 3.56DDD11 pKa = 3.64YY12 pKa = 10.18YYY14 pKa = 11.14KK15 pKa = 10.91QIVDDD20 pKa = 3.19KK21 pKa = 10.37GNVGNVDDD29 pKa = 3.79QDDD32 pKa = 3.28TIDDD36 pKa = 3.61SGPQDDD42 pKa = 3.46TIDDD46 pKa = 3.28TGITEEE52 pKa = 4.07DD53 pKa = 3.74GLVADDD59 pKa = 5.62DD60 pKa = 5.19YY61 pKa = 9.79YY62 pKa = 11.4NDDD65 pKa = 3.17TLVVSGVLGVALLAGEEE82 pKa = 4.41VQVQLDDD89 pKa = 3.25GNWYYY94 pKa = 7.66ATIDDD99 pKa = 3.54TGTGWSFDDD108 pKa = 3.92SSTTLDDD115 pKa = 3.43EE116 pKa = 4.77DDD118 pKa = 3.84HH119 pKa = 5.85FNAKKK124 pKa = 9.09VNAAGNDDD132 pKa = 3.6ATTSKKK138 pKa = 9.85VTVDDD143 pKa = 3.32TADDD147 pKa = 3.51AVAEEE152 pKa = 4.41TSIADDD158 pKa = 3.37DD159 pKa = 3.65GMVTDDD165 pKa = 5.62DD166 pKa = 4.25VTNDDD171 pKa = 3.27TLTVYYY177 pKa = 8.7TLDDD181 pKa = 3.12KK182 pKa = 11.31LGEEE186 pKa = 4.03SYYY189 pKa = 10.57EEE191 pKa = 3.6WGARR195 pKa = 11.84EEE197 pKa = 3.54QTPVKKK203 pKa = 10.18GVATVDDD210 pKa = 3.42TGTGWSFDDD219 pKa = 3.59SGNPHHH225 pKa = 6.77EE226 pKa = 5.41DD227 pKa = 3.14EEE229 pKa = 4.5YY230 pKa = 9.33YYY232 pKa = 10.1YYY234 pKa = 8.5EE235 pKa = 5.6RR236 pKa = 11.84VFDDD240 pKa = 3.49GGNSGSVDDD249 pKa = 3.52KKK251 pKa = 11.22VQIDDD256 pKa = 3.64VGPEEE261 pKa = 4.09LNLDDD266 pKa = 3.3TFEEE270 pKa = 4.34LTDDD274 pKa = 4.02YYY276 pKa = 11.0HH277 pKa = 7.68LDDD280 pKa = 5.06DDD282 pKa = 4.43EEE284 pKa = 4.71DD285 pKa = 4.45VSGINNDDD293 pKa = 4.54ITRR296 pKa = 11.84DDD298 pKa = 3.72YY299 pKa = 10.76ITIAGEEE306 pKa = 4.04SSSLQAGEEE315 pKa = 4.26YY316 pKa = 10.09QISTDDD322 pKa = 3.58EEE324 pKa = 3.98WEEE327 pKa = 4.67LLQDDD332 pKa = 4.79GKKK335 pKa = 9.08WSWKKK340 pKa = 10.11VDDD343 pKa = 3.16DDD345 pKa = 4.07YY346 pKa = 11.56VDDD349 pKa = 3.55TTTYYY354 pKa = 11.4YY355 pKa = 10.92RR356 pKa = 11.84LVDDD360 pKa = 3.85AGNEEE365 pKa = 4.04TGTVFKKK372 pKa = 10.29DDD374 pKa = 2.54GYYY377 pKa = 10.17EE378 pKa = 4.12TVDDD382 pKa = 3.62TAPDDD387 pKa = 5.56ILVAPGGVPEEE398 pKa = 4.34TSTTEEE404 pKa = 3.49EE405 pKa = 4.06TFDDD409 pKa = 3.13NTYYY413 pKa = 10.17KKK415 pKa = 10.19EEE417 pKa = 4.21NCYYY421 pKa = 8.1ALYYY425 pKa = 10.74DDD427 pKa = 3.59ANDDD431 pKa = 3.56SYYY434 pKa = 10.93LGVDDD439 pKa = 3.62AIHHH443 pKa = 5.38YY444 pKa = 7.9QADDD448 pKa = 3.38SGNWSMTTKKK458 pKa = 10.81PAGSNNLTFAVWDDD472 pKa = 3.68EE473 pKa = 4.17GNRR476 pKa = 11.84SQFGPSTSVGVSEEE490 pKa = 4.58TGSEEE495 pKa = 4.7DDD497 pKa = 3.45LTWGGTVNPGLGLNSAAVTINEEE520 pKa = 4.32DD521 pKa = 3.76LWSFFQSSAALSGTTTANAGWVFAAKKK548 pKa = 10.44QDDD551 pKa = 3.76DD552 pKa = 4.06YY553 pKa = 11.98ATYYY557 pKa = 10.77EEE559 pKa = 4.22PRR561 pKa = 11.84DDD563 pKa = 3.82TSAAYYY569 pKa = 9.85EE570 pKa = 4.11FVNAALFADDD580 pKa = 4.24YY581 pKa = 11.14RR582 pKa = 11.84DDD584 pKa = 2.95YYY586 pKa = 11.63DDD588 pKa = 4.08ISQVSSYYY596 pKa = 11.46NASRR600 pKa = 11.84TAMWTNNGDDD610 pKa = 3.43TYYY613 pKa = 9.91PGSINQGTLNHHH625 pKa = 6.72GGAIAYYY632 pKa = 9.03DD633 pKa = 3.84KK634 pKa = 11.43EE635 pKa = 4.33DDD637 pKa = 3.72YYY639 pKa = 11.47DDD641 pKa = 3.96VLADDD646 pKa = 3.86EEE648 pKa = 4.4DDD650 pKa = 3.32LSFLKKK656 pKa = 10.97VDDD659 pKa = 3.81DD660 pKa = 4.75SGGRR664 pKa = 11.84KKK666 pKa = 7.17TYYY669 pKa = 8.53EE670 pKa = 4.14NAGYYY675 pKa = 10.84YY676 pKa = 9.95GVPAGVDDD684 pKa = 2.92YYY686 pKa = 11.04SILHHH691 pKa = 5.75EE692 pKa = 4.38SAVDDD697 pKa = 4.4DDD699 pKa = 3.93DDD701 pKa = 3.84SVDDD705 pKa = 3.38AAHHH709 pKa = 5.36DDD711 pKa = 3.18LGGGNPGRR719 pKa = 11.84VLTVFHHH726 pKa = 7.07EEE728 pKa = 3.94NDDD731 pKa = 3.59SSQQFTMQDDD741 pKa = 3.32YY742 pKa = 9.91NVFRR746 pKa = 11.84NDDD749 pKa = 3.21YYY751 pKa = 11.61DDD753 pKa = 3.79YY754 pKa = 11.45NLSYYY759 pKa = 11.81MTWGDDD765 pKa = 3.75YY766 pKa = 11.2DD767 pKa = 4.22DDD769 pKa = 4.22YYY771 pKa = 11.66DDD773 pKa = 4.19FLSRR777 pKa = 11.84GNIKKK782 pKa = 10.67GVGNSDDD789 pKa = 3.29EE790 pKa = 4.19RR791 pKa = 11.84IYYY794 pKa = 10.92NDDD797 pKa = 3.61SGGLKKK803 pKa = 9.12ATEEE807 pKa = 4.48DD808 pKa = 3.7TWMGDDD814 pKa = 2.87LAGGTSFAVDDD825 pKa = 3.49NHHH828 pKa = 7.16DD829 pKa = 3.31LMDDD833 pKa = 4.4VEEE836 pKa = 4.8PNSRR840 pKa = 11.84AVQFGPTLYYY850 pKa = 10.68NQGDDD855 pKa = 4.49TWEEE859 pKa = 3.81MTMTEEE865 pKa = 4.29LGSTITGGVALDDD878 pKa = 4.13YY879 pKa = 11.05WDDD882 pKa = 3.46AMDDD886 pKa = 4.42VLYYY890 pKa = 10.18PSSTSLQSGADDD902 pKa = 3.71NTAPSKKK909 pKa = 10.61VNNTNIAADDD919 pKa = 3.94TSLQLRR925 pKa = 11.84ILDDD929 pKa = 3.77EEE931 pKa = 4.32MNVYYY936 pKa = 10.3YY937 pKa = 10.98NTVKKK942 pKa = 10.82YYY944 pKa = 10.73DD945 pKa = 3.53AGNCVATTLMNPQGSGSSNAMGLVSFYYY973 pKa = 11.33LDDD976 pKa = 3.42DDD978 pKa = 3.97EE979 pKa = 5.01YYY981 pKa = 11.03VQLLRR986 pKa = 11.84TTNGEEE992 pKa = 3.91DDD994 pKa = 3.23HH995 pKa = 6.62GSVASQGGFTNSTVNSSWGGLTTGKKK1021 pKa = 9.85HHH1023 pKa = 7.78DD1024 pKa = 3.68YYY1026 pKa = 10.05LVGEEE1031 pKa = 4.77DDD1033 pKa = 3.68STASSTNEEE1042 pKa = 3.3GIVGSGYYY1050 pKa = 10.46DDD1052 pKa = 3.53FVSSAGDDD1060 pKa = 3.75DD1061 pKa = 3.32FNGSGGWSQLVISDDD1076 pKa = 4.57VWEEE1080 pKa = 4.24DDD1082 pKa = 3.56GLDDD1086 pKa = 3.22VDDD1089 pKa = 3.43YY1090 pKa = 11.22KKK1092 pKa = 10.24TGGGVQANLATGVVTGAAGNDDD1114 pKa = 3.36LINIEEE1120 pKa = 4.19LLGTSADDD1128 pKa = 3.86DD1129 pKa = 3.74FTDDD1133 pKa = 3.43AANNVFEEE1141 pKa = 4.5RR1142 pKa = 11.84GGNDDD1147 pKa = 3.05YYY1149 pKa = 11.27LTGGGRR1155 pKa = 11.84DDD1157 pKa = 3.31LLYYY1161 pKa = 10.35EE1162 pKa = 4.23LADDD1166 pKa = 4.12DDD1168 pKa = 4.05TGGNGSDDD1176 pKa = 4.24INDDD1180 pKa = 3.56TVGLVKKK1187 pKa = 10.55DD1188 pKa = 3.85ADDD1191 pKa = 4.18DDD1193 pKa = 4.22INISDDD1199 pKa = 4.06LNYYY1203 pKa = 9.84GTVGTYYY1210 pKa = 8.8DDD1212 pKa = 3.21SGTYYY1217 pKa = 8.56HH1218 pKa = 7.83DDD1220 pKa = 3.42ASEEE1224 pKa = 4.2VNDDD1228 pKa = 4.24VKKK1231 pKa = 9.88TTSGGNTEEE1240 pKa = 3.91AIDDD1244 pKa = 3.61DDD1246 pKa = 3.5AGNYYY1251 pKa = 6.54TLVTLEEE1258 pKa = 4.04TEEE1261 pKa = 4.04DDD1263 pKa = 3.96LSLLTNQQLIVEEE1276 pKa = 4.64SADDD1280 pKa = 3.51GASGAAASQQEEE1292 pKa = 4.37DD1293 pKa = 4.24EEE1295 pKa = 4.68STGSTGGDDD1304 pKa = 3.72DD1305 pKa = 3.79TASDDD1310 pKa = 4.75STTAPIADDD1319 pKa = 3.83SGNTTDDD1326 pKa = 3.24SGVVDDD1332 pKa = 3.9ADDD1335 pKa = 3.25VTTRR1339 pKa = 11.84VEEE1342 pKa = 4.34DD1343 pKa = 4.67DD1344 pKa = 5.04VTQLNSPTVI
Molecular weight: 142.25 kDa
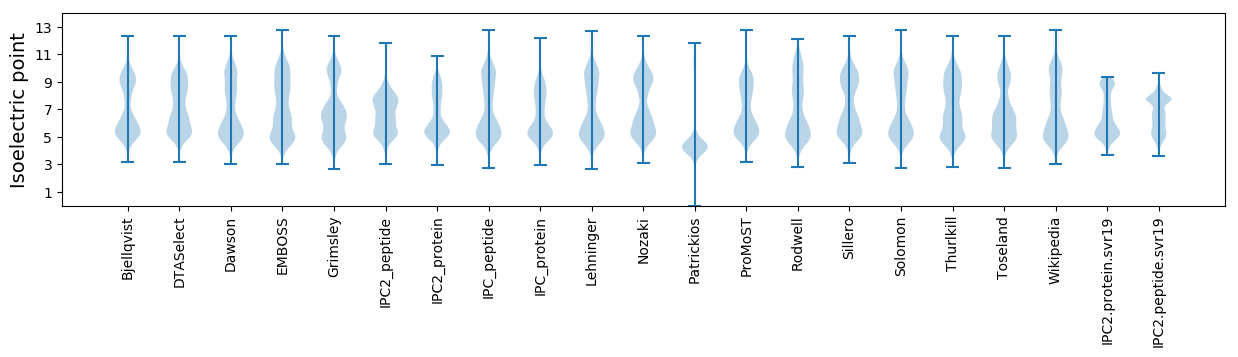
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H0J2U3|A0A1H0J2U3_9DELT Cu+-exporting ATPase OS=Desulforhopalus singaporensis OX=91360 GN=SAMN05660330_00122 PE=3 SV=1
MM1 pKa = 7.48AKK3 pKa = 9.86KK4 pKa = 9.04RR5 pKa = 11.84TYY7 pKa = 10.26RR8 pKa = 11.84VVDD11 pKa = 3.23GRR13 pKa = 11.84MNFVCPEE20 pKa = 3.78CHH22 pKa = 5.51TRR24 pKa = 11.84RR25 pKa = 11.84LVGVSPHH32 pKa = 6.09LKK34 pKa = 9.55NRR36 pKa = 11.84SIKK39 pKa = 9.26CHH41 pKa = 5.79KK42 pKa = 9.87CSTVVSCTLNRR53 pKa = 11.84RR54 pKa = 11.84EE55 pKa = 4.49TIRR58 pKa = 11.84EE59 pKa = 3.85QQFGKK64 pKa = 10.78VLLTRR69 pKa = 11.84YY70 pKa = 9.35DD71 pKa = 3.55RR72 pKa = 11.84SVVEE76 pKa = 4.64VNLLDD81 pKa = 3.49ISTWGVGFEE90 pKa = 4.37LPFRR94 pKa = 11.84EE95 pKa = 4.31LKK97 pKa = 10.28KK98 pKa = 10.79LSVGEE103 pKa = 4.26RR104 pKa = 11.84IEE106 pKa = 5.76FRR108 pKa = 11.84CPWNPKK114 pKa = 9.95LLGSSAYY121 pKa = 10.03VIRR124 pKa = 11.84AINGRR129 pKa = 11.84RR130 pKa = 11.84VGAEE134 pKa = 3.95RR135 pKa = 11.84LRR137 pKa = 11.84KK138 pKa = 9.45SS139 pKa = 3.13
MM1 pKa = 7.48AKK3 pKa = 9.86KK4 pKa = 9.04RR5 pKa = 11.84TYY7 pKa = 10.26RR8 pKa = 11.84VVDD11 pKa = 3.23GRR13 pKa = 11.84MNFVCPEE20 pKa = 3.78CHH22 pKa = 5.51TRR24 pKa = 11.84RR25 pKa = 11.84LVGVSPHH32 pKa = 6.09LKK34 pKa = 9.55NRR36 pKa = 11.84SIKK39 pKa = 9.26CHH41 pKa = 5.79KK42 pKa = 9.87CSTVVSCTLNRR53 pKa = 11.84RR54 pKa = 11.84EE55 pKa = 4.49TIRR58 pKa = 11.84EE59 pKa = 3.85QQFGKK64 pKa = 10.78VLLTRR69 pKa = 11.84YY70 pKa = 9.35DD71 pKa = 3.55RR72 pKa = 11.84SVVEE76 pKa = 4.64VNLLDD81 pKa = 3.49ISTWGVGFEE90 pKa = 4.37LPFRR94 pKa = 11.84EE95 pKa = 4.31LKK97 pKa = 10.28KK98 pKa = 10.79LSVGEE103 pKa = 4.26RR104 pKa = 11.84IEE106 pKa = 5.76FRR108 pKa = 11.84CPWNPKK114 pKa = 9.95LLGSSAYY121 pKa = 10.03VIRR124 pKa = 11.84AINGRR129 pKa = 11.84RR130 pKa = 11.84VGAEE134 pKa = 3.95RR135 pKa = 11.84LRR137 pKa = 11.84KK138 pKa = 9.45SS139 pKa = 3.13
Molecular weight: 16.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1432539 |
15 |
1863 |
329.5 |
36.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.161 ± 0.036 | 1.461 ± 0.017 |
5.435 ± 0.03 | 6.358 ± 0.034 |
4.284 ± 0.028 | 7.795 ± 0.037 |
2.031 ± 0.016 | 6.739 ± 0.031 |
5.615 ± 0.032 | 9.987 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.678 ± 0.017 | 3.759 ± 0.021 |
4.197 ± 0.022 | 3.362 ± 0.023 |
5.392 ± 0.035 | 6.067 ± 0.024 |
5.346 ± 0.025 | 7.229 ± 0.029 |
1.073 ± 0.014 | 3.032 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
