
Salinimonas sediminis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Salinimonas
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
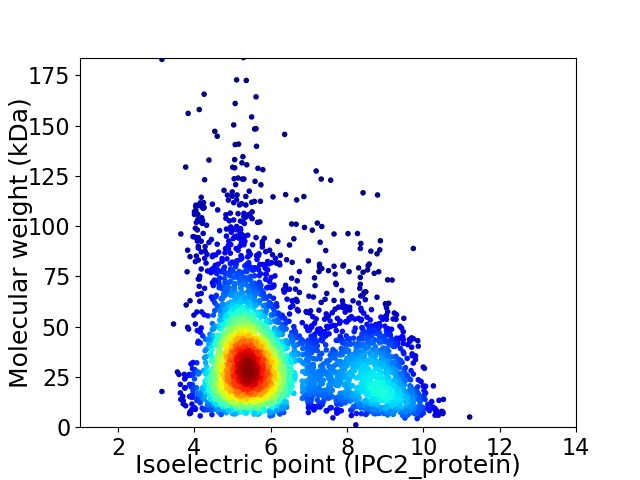
Virtual 2D-PAGE plot for 3700 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
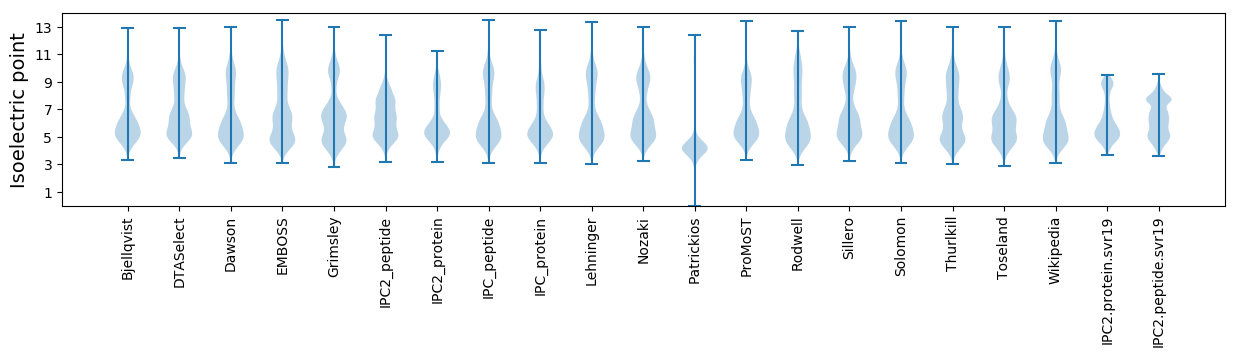
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346NHQ7|A0A346NHQ7_9ALTE Class A beta-lactamase-related serine hydrolase OS=Salinimonas sediminis OX=2303538 GN=D0Y50_00975 PE=4 SV=1
MM1 pKa = 7.7KK2 pKa = 9.24LTRR5 pKa = 11.84YY6 pKa = 9.52GVLGVMAGTVLAACGGGSDD25 pKa = 4.17NSSSSTQNGSQTAGKK40 pKa = 9.68DD41 pKa = 3.06ASQIIGNASAHH52 pKa = 6.01SVSEE56 pKa = 4.09LNKK59 pKa = 9.94AARR62 pKa = 11.84SLASASYY69 pKa = 9.67TGSQEE74 pKa = 3.99KK75 pKa = 11.13AEE77 pKa = 4.54LDD79 pKa = 3.55FLLAQQVTRR88 pKa = 11.84MLLDD92 pKa = 4.65DD93 pKa = 4.59DD94 pKa = 4.21VLRR97 pKa = 11.84VPEE100 pKa = 4.26IVTHH104 pKa = 5.76EE105 pKa = 4.4LSPDD109 pKa = 2.95ARR111 pKa = 11.84QDD113 pKa = 3.44EE114 pKa = 4.93NINATFDD121 pKa = 3.79CDD123 pKa = 3.56EE124 pKa = 5.14GGSASYY130 pKa = 10.45SGQLNEE136 pKa = 4.66SGEE139 pKa = 4.29ASLSLNFDD147 pKa = 3.47NCKK150 pKa = 10.43SYY152 pKa = 11.79YY153 pKa = 9.96NDD155 pKa = 3.43SALTGVAALVIKK167 pKa = 10.84LDD169 pKa = 3.67VQQNTSAYY177 pKa = 8.74TLYY180 pKa = 10.62FDD182 pKa = 4.33NLRR185 pKa = 11.84WHH187 pKa = 7.45DD188 pKa = 3.85GQAVSLSGLMAFNEE202 pKa = 4.37SADD205 pKa = 3.05WEE207 pKa = 4.52MPYY210 pKa = 7.92TTLSQQYY217 pKa = 9.3IALSVGNEE225 pKa = 4.07TYY227 pKa = 11.18LLDD230 pKa = 4.92AEE232 pKa = 4.74TTGEE236 pKa = 4.14AEE238 pKa = 4.19SGSLLEE244 pKa = 4.3SMQGRR249 pKa = 11.84LSVASRR255 pKa = 11.84GYY257 pKa = 10.06IDD259 pKa = 5.87FSYY262 pKa = 10.87EE263 pKa = 3.98PQTYY267 pKa = 8.35TYY269 pKa = 10.32DD270 pKa = 3.82FNGSTLFFNGNEE282 pKa = 3.9AYY284 pKa = 11.09VNFEE288 pKa = 3.46QDD290 pKa = 2.47TAAFVIDD297 pKa = 3.83TDD299 pKa = 4.22QDD301 pKa = 3.48GTTDD305 pKa = 3.06AGKK308 pKa = 10.03ILSLADD314 pKa = 4.17FFAPPPADD322 pKa = 3.47ASLNPLTEE330 pKa = 4.28ISSQPYY336 pKa = 9.91AGEE339 pKa = 4.5PYY341 pKa = 10.18TNEE344 pKa = 3.36SYY346 pKa = 9.61YY347 pKa = 10.62TVSTDD352 pKa = 2.79IDD354 pKa = 3.34ISPGYY359 pKa = 9.97FFDD362 pKa = 4.33IEE364 pKa = 4.51TPRR367 pKa = 11.84SQLEE371 pKa = 3.75ISYY374 pKa = 9.88NWYY377 pKa = 9.92INGILLEE384 pKa = 4.47GVHH387 pKa = 6.7GSTLPAYY394 pKa = 10.09SAGVHH399 pKa = 6.72DD400 pKa = 5.23EE401 pKa = 4.42VTVSMVVSDD410 pKa = 3.69ATHH413 pKa = 6.55SYY415 pKa = 10.87EE416 pKa = 5.78SYY418 pKa = 10.04QQYY421 pKa = 10.08IWLEE425 pKa = 4.0DD426 pKa = 3.55MPAEE430 pKa = 4.09LTVTDD435 pKa = 4.51MPASIAAGEE444 pKa = 4.32TVSFSVVLTDD454 pKa = 4.97PDD456 pKa = 3.71IEE458 pKa = 4.51SADD461 pKa = 3.47NLARR465 pKa = 11.84LTNAPQGASMSEE477 pKa = 4.22DD478 pKa = 3.19GTVTWQVPQDD488 pKa = 3.92LLFSEE493 pKa = 4.2QNYY496 pKa = 9.19YY497 pKa = 10.88FGFSNRR503 pKa = 11.84LNPNEE508 pKa = 4.33TIEE511 pKa = 4.25KK512 pKa = 9.34TVTATVNASQPLPLFRR528 pKa = 11.84SGIEE532 pKa = 3.7VPAANRR538 pKa = 11.84SQWVVDD544 pKa = 3.44IDD546 pKa = 4.51GDD548 pKa = 4.16GQNEE552 pKa = 4.3LLSTDD557 pKa = 2.88NDD559 pKa = 3.22QRR561 pKa = 11.84VFLLSLDD568 pKa = 3.51NGQYY572 pKa = 6.43QQKK575 pKa = 8.31WAYY578 pKa = 9.36PYY580 pKa = 10.93VLSQQEE586 pKa = 4.38TIQQVLPIGRR596 pKa = 11.84NQAGSKK602 pKa = 9.68PILVVTDD609 pKa = 3.57HH610 pKa = 6.77ALWRR614 pKa = 11.84IDD616 pKa = 5.49DD617 pKa = 4.4LAQPARR623 pKa = 11.84QIFNTDD629 pKa = 2.56NFLRR633 pKa = 11.84SALLADD639 pKa = 3.56IDD641 pKa = 4.21GDD643 pKa = 3.93GTEE646 pKa = 3.94EE647 pKa = 4.91LIYY650 pKa = 10.62HH651 pKa = 7.1AAADD655 pKa = 4.28AYY657 pKa = 10.63SATLAIVNVVALDD670 pKa = 4.08DD671 pKa = 4.53PSTEE675 pKa = 3.96LMSFTVDD682 pKa = 2.88EE683 pKa = 5.02FSTFDD688 pKa = 3.39VGNVDD693 pKa = 2.88TDD695 pKa = 3.81ANLEE699 pKa = 4.16LVLNTGSVFDD709 pKa = 5.02LVTAEE714 pKa = 4.82NEE716 pKa = 4.09WQLGTGFSTQYY727 pKa = 9.32VTTGDD732 pKa = 3.9LNGDD736 pKa = 4.14GIDD739 pKa = 4.32EE740 pKa = 5.0IISADD745 pKa = 3.18SWGAMYY751 pKa = 10.66VFSVTEE757 pKa = 3.98KK758 pKa = 9.28TQLDD762 pKa = 3.68SRR764 pKa = 11.84DD765 pKa = 3.77AEE767 pKa = 4.74DD768 pKa = 3.58ICDD771 pKa = 3.66LRR773 pKa = 11.84AFNLNQDD780 pKa = 3.93DD781 pKa = 3.87TDD783 pKa = 4.25EE784 pKa = 5.69IIISDD789 pKa = 4.26CQWGEE794 pKa = 4.0VQAFTLDD801 pKa = 3.65EE802 pKa = 4.32NSALAQLWSVDD813 pKa = 3.91LQEE816 pKa = 5.23HH817 pKa = 6.74GSTSLSAGDD826 pKa = 4.12IDD828 pKa = 4.26NDD830 pKa = 3.59GHH832 pKa = 8.37IEE834 pKa = 4.08VLWGSGVSSSGRR846 pKa = 11.84DD847 pKa = 3.2ILVTADD853 pKa = 3.55VVGTTATVKK862 pKa = 10.72PEE864 pKa = 4.0TSPVQLDD871 pKa = 3.58AFAAAGWATTGADD884 pKa = 3.58DD885 pKa = 3.87EE886 pKa = 4.66QGVFIVPTTDD896 pKa = 2.54SGYY899 pKa = 10.65SGLSIARR906 pKa = 11.84LNQNGTVNVNDD917 pKa = 4.18KK918 pKa = 10.78FSSFWQSGMFSAITDD933 pKa = 3.65TDD935 pKa = 3.27NDD937 pKa = 4.94GIDD940 pKa = 5.52DD941 pKa = 4.05ILLPTTGTYY950 pKa = 10.23DD951 pKa = 3.35YY952 pKa = 11.36GLGAFALSDD961 pKa = 4.18LSSLWTVYY969 pKa = 10.97GDD971 pKa = 3.43YY972 pKa = 9.49DD973 pKa = 4.13TNINQIKK980 pKa = 9.88LQDD983 pKa = 3.71INNDD987 pKa = 2.6GHH989 pKa = 6.21TDD991 pKa = 3.18VMFADD996 pKa = 4.34GDD998 pKa = 4.05VLTVIDD1004 pKa = 4.39TQTQEE1009 pKa = 3.61QLTSYY1014 pKa = 10.52RR1015 pKa = 11.84FSQSIADD1022 pKa = 4.11FDD1024 pKa = 3.95VADD1027 pKa = 3.78IDD1029 pKa = 4.24GVTTVLVTLRR1039 pKa = 11.84EE1040 pKa = 4.03QVHH1043 pKa = 6.16ILTLKK1048 pKa = 7.09EE1049 pKa = 3.89TRR1051 pKa = 11.84FTVVDD1056 pKa = 4.0SVSDD1060 pKa = 3.39ILCNRR1065 pKa = 11.84ILFYY1069 pKa = 10.83GQSNQRR1075 pKa = 11.84SVACLWTDD1083 pKa = 3.17YY1084 pKa = 10.81GASLRR1089 pKa = 11.84TMRR1092 pKa = 11.84IVDD1095 pKa = 3.59NQATEE1100 pKa = 4.39LEE1102 pKa = 4.68TFALEE1107 pKa = 4.13HH1108 pKa = 6.1NAVAIAVDD1116 pKa = 3.57PTDD1119 pKa = 3.53PQRR1122 pKa = 11.84FIIAANTEE1130 pKa = 4.27DD1131 pKa = 2.97GWYY1134 pKa = 9.88DD1135 pKa = 3.42ARR1137 pKa = 11.84SQLHH1141 pKa = 6.41SLTAQGKK1148 pKa = 7.73LVWSGPVLIGSANVASLQLRR1168 pKa = 11.84EE1169 pKa = 4.06QTSGKK1174 pKa = 10.6LEE1176 pKa = 3.91ILYY1179 pKa = 7.92GTSKK1183 pKa = 10.6GMYY1186 pKa = 8.4WIHH1189 pKa = 6.63
MM1 pKa = 7.7KK2 pKa = 9.24LTRR5 pKa = 11.84YY6 pKa = 9.52GVLGVMAGTVLAACGGGSDD25 pKa = 4.17NSSSSTQNGSQTAGKK40 pKa = 9.68DD41 pKa = 3.06ASQIIGNASAHH52 pKa = 6.01SVSEE56 pKa = 4.09LNKK59 pKa = 9.94AARR62 pKa = 11.84SLASASYY69 pKa = 9.67TGSQEE74 pKa = 3.99KK75 pKa = 11.13AEE77 pKa = 4.54LDD79 pKa = 3.55FLLAQQVTRR88 pKa = 11.84MLLDD92 pKa = 4.65DD93 pKa = 4.59DD94 pKa = 4.21VLRR97 pKa = 11.84VPEE100 pKa = 4.26IVTHH104 pKa = 5.76EE105 pKa = 4.4LSPDD109 pKa = 2.95ARR111 pKa = 11.84QDD113 pKa = 3.44EE114 pKa = 4.93NINATFDD121 pKa = 3.79CDD123 pKa = 3.56EE124 pKa = 5.14GGSASYY130 pKa = 10.45SGQLNEE136 pKa = 4.66SGEE139 pKa = 4.29ASLSLNFDD147 pKa = 3.47NCKK150 pKa = 10.43SYY152 pKa = 11.79YY153 pKa = 9.96NDD155 pKa = 3.43SALTGVAALVIKK167 pKa = 10.84LDD169 pKa = 3.67VQQNTSAYY177 pKa = 8.74TLYY180 pKa = 10.62FDD182 pKa = 4.33NLRR185 pKa = 11.84WHH187 pKa = 7.45DD188 pKa = 3.85GQAVSLSGLMAFNEE202 pKa = 4.37SADD205 pKa = 3.05WEE207 pKa = 4.52MPYY210 pKa = 7.92TTLSQQYY217 pKa = 9.3IALSVGNEE225 pKa = 4.07TYY227 pKa = 11.18LLDD230 pKa = 4.92AEE232 pKa = 4.74TTGEE236 pKa = 4.14AEE238 pKa = 4.19SGSLLEE244 pKa = 4.3SMQGRR249 pKa = 11.84LSVASRR255 pKa = 11.84GYY257 pKa = 10.06IDD259 pKa = 5.87FSYY262 pKa = 10.87EE263 pKa = 3.98PQTYY267 pKa = 8.35TYY269 pKa = 10.32DD270 pKa = 3.82FNGSTLFFNGNEE282 pKa = 3.9AYY284 pKa = 11.09VNFEE288 pKa = 3.46QDD290 pKa = 2.47TAAFVIDD297 pKa = 3.83TDD299 pKa = 4.22QDD301 pKa = 3.48GTTDD305 pKa = 3.06AGKK308 pKa = 10.03ILSLADD314 pKa = 4.17FFAPPPADD322 pKa = 3.47ASLNPLTEE330 pKa = 4.28ISSQPYY336 pKa = 9.91AGEE339 pKa = 4.5PYY341 pKa = 10.18TNEE344 pKa = 3.36SYY346 pKa = 9.61YY347 pKa = 10.62TVSTDD352 pKa = 2.79IDD354 pKa = 3.34ISPGYY359 pKa = 9.97FFDD362 pKa = 4.33IEE364 pKa = 4.51TPRR367 pKa = 11.84SQLEE371 pKa = 3.75ISYY374 pKa = 9.88NWYY377 pKa = 9.92INGILLEE384 pKa = 4.47GVHH387 pKa = 6.7GSTLPAYY394 pKa = 10.09SAGVHH399 pKa = 6.72DD400 pKa = 5.23EE401 pKa = 4.42VTVSMVVSDD410 pKa = 3.69ATHH413 pKa = 6.55SYY415 pKa = 10.87EE416 pKa = 5.78SYY418 pKa = 10.04QQYY421 pKa = 10.08IWLEE425 pKa = 4.0DD426 pKa = 3.55MPAEE430 pKa = 4.09LTVTDD435 pKa = 4.51MPASIAAGEE444 pKa = 4.32TVSFSVVLTDD454 pKa = 4.97PDD456 pKa = 3.71IEE458 pKa = 4.51SADD461 pKa = 3.47NLARR465 pKa = 11.84LTNAPQGASMSEE477 pKa = 4.22DD478 pKa = 3.19GTVTWQVPQDD488 pKa = 3.92LLFSEE493 pKa = 4.2QNYY496 pKa = 9.19YY497 pKa = 10.88FGFSNRR503 pKa = 11.84LNPNEE508 pKa = 4.33TIEE511 pKa = 4.25KK512 pKa = 9.34TVTATVNASQPLPLFRR528 pKa = 11.84SGIEE532 pKa = 3.7VPAANRR538 pKa = 11.84SQWVVDD544 pKa = 3.44IDD546 pKa = 4.51GDD548 pKa = 4.16GQNEE552 pKa = 4.3LLSTDD557 pKa = 2.88NDD559 pKa = 3.22QRR561 pKa = 11.84VFLLSLDD568 pKa = 3.51NGQYY572 pKa = 6.43QQKK575 pKa = 8.31WAYY578 pKa = 9.36PYY580 pKa = 10.93VLSQQEE586 pKa = 4.38TIQQVLPIGRR596 pKa = 11.84NQAGSKK602 pKa = 9.68PILVVTDD609 pKa = 3.57HH610 pKa = 6.77ALWRR614 pKa = 11.84IDD616 pKa = 5.49DD617 pKa = 4.4LAQPARR623 pKa = 11.84QIFNTDD629 pKa = 2.56NFLRR633 pKa = 11.84SALLADD639 pKa = 3.56IDD641 pKa = 4.21GDD643 pKa = 3.93GTEE646 pKa = 3.94EE647 pKa = 4.91LIYY650 pKa = 10.62HH651 pKa = 7.1AAADD655 pKa = 4.28AYY657 pKa = 10.63SATLAIVNVVALDD670 pKa = 4.08DD671 pKa = 4.53PSTEE675 pKa = 3.96LMSFTVDD682 pKa = 2.88EE683 pKa = 5.02FSTFDD688 pKa = 3.39VGNVDD693 pKa = 2.88TDD695 pKa = 3.81ANLEE699 pKa = 4.16LVLNTGSVFDD709 pKa = 5.02LVTAEE714 pKa = 4.82NEE716 pKa = 4.09WQLGTGFSTQYY727 pKa = 9.32VTTGDD732 pKa = 3.9LNGDD736 pKa = 4.14GIDD739 pKa = 4.32EE740 pKa = 5.0IISADD745 pKa = 3.18SWGAMYY751 pKa = 10.66VFSVTEE757 pKa = 3.98KK758 pKa = 9.28TQLDD762 pKa = 3.68SRR764 pKa = 11.84DD765 pKa = 3.77AEE767 pKa = 4.74DD768 pKa = 3.58ICDD771 pKa = 3.66LRR773 pKa = 11.84AFNLNQDD780 pKa = 3.93DD781 pKa = 3.87TDD783 pKa = 4.25EE784 pKa = 5.69IIISDD789 pKa = 4.26CQWGEE794 pKa = 4.0VQAFTLDD801 pKa = 3.65EE802 pKa = 4.32NSALAQLWSVDD813 pKa = 3.91LQEE816 pKa = 5.23HH817 pKa = 6.74GSTSLSAGDD826 pKa = 4.12IDD828 pKa = 4.26NDD830 pKa = 3.59GHH832 pKa = 8.37IEE834 pKa = 4.08VLWGSGVSSSGRR846 pKa = 11.84DD847 pKa = 3.2ILVTADD853 pKa = 3.55VVGTTATVKK862 pKa = 10.72PEE864 pKa = 4.0TSPVQLDD871 pKa = 3.58AFAAAGWATTGADD884 pKa = 3.58DD885 pKa = 3.87EE886 pKa = 4.66QGVFIVPTTDD896 pKa = 2.54SGYY899 pKa = 10.65SGLSIARR906 pKa = 11.84LNQNGTVNVNDD917 pKa = 4.18KK918 pKa = 10.78FSSFWQSGMFSAITDD933 pKa = 3.65TDD935 pKa = 3.27NDD937 pKa = 4.94GIDD940 pKa = 5.52DD941 pKa = 4.05ILLPTTGTYY950 pKa = 10.23DD951 pKa = 3.35YY952 pKa = 11.36GLGAFALSDD961 pKa = 4.18LSSLWTVYY969 pKa = 10.97GDD971 pKa = 3.43YY972 pKa = 9.49DD973 pKa = 4.13TNINQIKK980 pKa = 9.88LQDD983 pKa = 3.71INNDD987 pKa = 2.6GHH989 pKa = 6.21TDD991 pKa = 3.18VMFADD996 pKa = 4.34GDD998 pKa = 4.05VLTVIDD1004 pKa = 4.39TQTQEE1009 pKa = 3.61QLTSYY1014 pKa = 10.52RR1015 pKa = 11.84FSQSIADD1022 pKa = 4.11FDD1024 pKa = 3.95VADD1027 pKa = 3.78IDD1029 pKa = 4.24GVTTVLVTLRR1039 pKa = 11.84EE1040 pKa = 4.03QVHH1043 pKa = 6.16ILTLKK1048 pKa = 7.09EE1049 pKa = 3.89TRR1051 pKa = 11.84FTVVDD1056 pKa = 4.0SVSDD1060 pKa = 3.39ILCNRR1065 pKa = 11.84ILFYY1069 pKa = 10.83GQSNQRR1075 pKa = 11.84SVACLWTDD1083 pKa = 3.17YY1084 pKa = 10.81GASLRR1089 pKa = 11.84TMRR1092 pKa = 11.84IVDD1095 pKa = 3.59NQATEE1100 pKa = 4.39LEE1102 pKa = 4.68TFALEE1107 pKa = 4.13HH1108 pKa = 6.1NAVAIAVDD1116 pKa = 3.57PTDD1119 pKa = 3.53PQRR1122 pKa = 11.84FIIAANTEE1130 pKa = 4.27DD1131 pKa = 2.97GWYY1134 pKa = 9.88DD1135 pKa = 3.42ARR1137 pKa = 11.84SQLHH1141 pKa = 6.41SLTAQGKK1148 pKa = 7.73LVWSGPVLIGSANVASLQLRR1168 pKa = 11.84EE1169 pKa = 4.06QTSGKK1174 pKa = 10.6LEE1176 pKa = 3.91ILYY1179 pKa = 7.92GTSKK1183 pKa = 10.6GMYY1186 pKa = 8.4WIHH1189 pKa = 6.63
Molecular weight: 129.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346NH93|A0A346NH93_9ALTE Probable chromosome-partitioning protein ParB OS=Salinimonas sediminis OX=2303538 GN=D0Y50_00070 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1236480 |
11 |
1702 |
334.2 |
36.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.838 ± 0.047 | 1.005 ± 0.015 |
5.599 ± 0.038 | 5.609 ± 0.041 |
4.018 ± 0.024 | 6.89 ± 0.038 |
2.41 ± 0.023 | 5.817 ± 0.03 |
4.379 ± 0.032 | 10.259 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.526 ± 0.023 | 3.993 ± 0.023 |
4.206 ± 0.026 | 5.03 ± 0.034 |
4.984 ± 0.028 | 6.27 ± 0.033 |
5.732 ± 0.027 | 6.989 ± 0.033 |
1.287 ± 0.017 | 3.159 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
