
Devosia sp. DBB001
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Devosiaceae; Devosia; unclassified Devosia
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
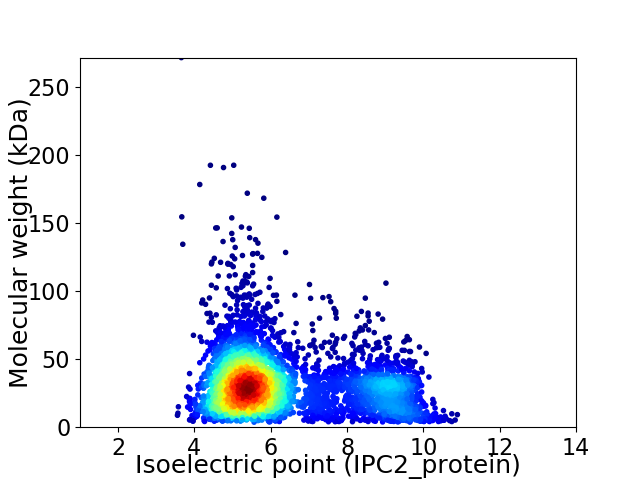
Virtual 2D-PAGE plot for 4502 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
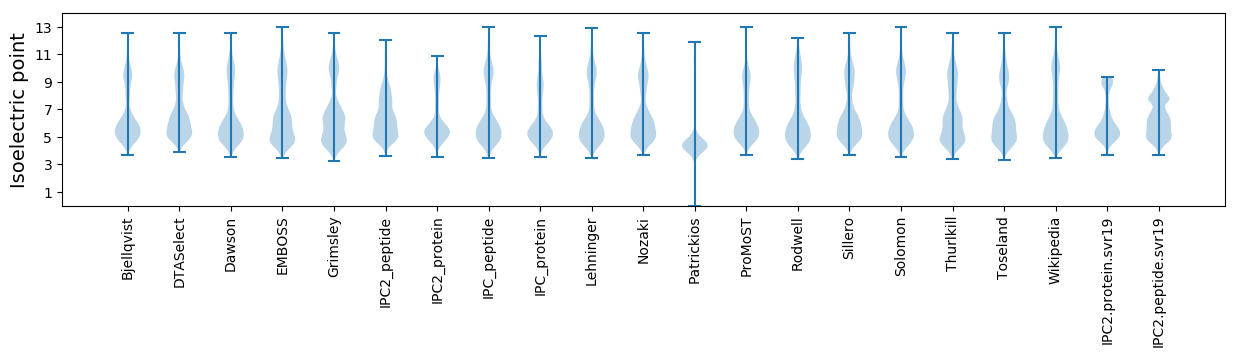
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A024K8I7|A0A024K8I7_9RHIZ CblY a non-orthologous displasment for Alpha-rib azole-5'-phosphate phosphatase OS=Devosia sp. DBB001 OX=1380412 PE=4 SV=1
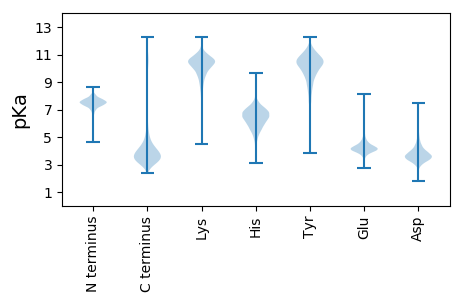
MM1 pKa = 8.02AIGRR5 pKa = 11.84LLLNSRR11 pKa = 11.84LGFALIGALLCSPFISIEE29 pKa = 3.63PAYY32 pKa = 10.74ADD34 pKa = 4.18LRR36 pKa = 11.84VCNEE40 pKa = 3.77TGNQVSIALGYY51 pKa = 9.19RR52 pKa = 11.84AEE54 pKa = 4.78RR55 pKa = 11.84GWQSEE60 pKa = 4.61GWWVATPQQCAVVYY74 pKa = 10.65QGDD77 pKa = 4.02LNSRR81 pKa = 11.84FYY83 pKa = 11.29YY84 pKa = 10.45LYY86 pKa = 10.6VADD89 pKa = 5.84DD90 pKa = 3.61IGGGAWDD97 pKa = 4.22GSVYY101 pKa = 9.99MCTRR105 pKa = 11.84DD106 pKa = 3.19EE107 pKa = 4.38SFTIFGVEE115 pKa = 3.75DD116 pKa = 3.75CLARR120 pKa = 11.84GYY122 pKa = 10.83EE123 pKa = 3.73RR124 pKa = 11.84TGFFEE129 pKa = 4.99VDD131 pKa = 3.17TQNRR135 pKa = 11.84TDD137 pKa = 2.99WTLQLTEE144 pKa = 4.43NRR146 pKa = 11.84DD147 pKa = 4.22AIPEE151 pKa = 4.23GDD153 pKa = 4.12DD154 pKa = 3.41ATGEE158 pKa = 4.11PLGDD162 pKa = 3.63EE163 pKa = 4.51TGGVPPDD170 pKa = 3.84QQDD173 pKa = 3.55PPPPDD178 pKa = 3.44EE179 pKa = 4.27TTPAPEE185 pKa = 4.6GPAAIQGSDD194 pKa = 2.77
MM1 pKa = 8.02AIGRR5 pKa = 11.84LLLNSRR11 pKa = 11.84LGFALIGALLCSPFISIEE29 pKa = 3.63PAYY32 pKa = 10.74ADD34 pKa = 4.18LRR36 pKa = 11.84VCNEE40 pKa = 3.77TGNQVSIALGYY51 pKa = 9.19RR52 pKa = 11.84AEE54 pKa = 4.78RR55 pKa = 11.84GWQSEE60 pKa = 4.61GWWVATPQQCAVVYY74 pKa = 10.65QGDD77 pKa = 4.02LNSRR81 pKa = 11.84FYY83 pKa = 11.29YY84 pKa = 10.45LYY86 pKa = 10.6VADD89 pKa = 5.84DD90 pKa = 3.61IGGGAWDD97 pKa = 4.22GSVYY101 pKa = 9.99MCTRR105 pKa = 11.84DD106 pKa = 3.19EE107 pKa = 4.38SFTIFGVEE115 pKa = 3.75DD116 pKa = 3.75CLARR120 pKa = 11.84GYY122 pKa = 10.83EE123 pKa = 3.73RR124 pKa = 11.84TGFFEE129 pKa = 4.99VDD131 pKa = 3.17TQNRR135 pKa = 11.84TDD137 pKa = 2.99WTLQLTEE144 pKa = 4.43NRR146 pKa = 11.84DD147 pKa = 4.22AIPEE151 pKa = 4.23GDD153 pKa = 4.12DD154 pKa = 3.41ATGEE158 pKa = 4.11PLGDD162 pKa = 3.63EE163 pKa = 4.51TGGVPPDD170 pKa = 3.84QQDD173 pKa = 3.55PPPPDD178 pKa = 3.44EE179 pKa = 4.27TTPAPEE185 pKa = 4.6GPAAIQGSDD194 pKa = 2.77
Molecular weight: 21.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A024KHT6|A0A024KHT6_9RHIZ Putative diheme cytochrome c-553 OS=Devosia sp. DBB001 OX=1380412 PE=4 SV=1
MM1 pKa = 7.43RR2 pKa = 11.84CARR5 pKa = 11.84RR6 pKa = 11.84ATDD9 pKa = 3.25RR10 pKa = 11.84TSRR13 pKa = 11.84ATRR16 pKa = 11.84RR17 pKa = 11.84GVRR20 pKa = 11.84TTRR23 pKa = 11.84RR24 pKa = 11.84TGRR27 pKa = 11.84IIHH30 pKa = 6.34RR31 pKa = 11.84AARR34 pKa = 11.84AAPWAHH40 pKa = 4.59TTSRR44 pKa = 11.84AAHH47 pKa = 5.3TARR50 pKa = 11.84RR51 pKa = 11.84ARR53 pKa = 11.84TTSRR57 pKa = 11.84TTRR60 pKa = 11.84ATHH63 pKa = 6.68RR64 pKa = 11.84ARR66 pKa = 11.84HH67 pKa = 4.56ICRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84NAACLCGRR80 pKa = 11.84AGIGSRR86 pKa = 11.84LL87 pKa = 3.42
MM1 pKa = 7.43RR2 pKa = 11.84CARR5 pKa = 11.84RR6 pKa = 11.84ATDD9 pKa = 3.25RR10 pKa = 11.84TSRR13 pKa = 11.84ATRR16 pKa = 11.84RR17 pKa = 11.84GVRR20 pKa = 11.84TTRR23 pKa = 11.84RR24 pKa = 11.84TGRR27 pKa = 11.84IIHH30 pKa = 6.34RR31 pKa = 11.84AARR34 pKa = 11.84AAPWAHH40 pKa = 4.59TTSRR44 pKa = 11.84AAHH47 pKa = 5.3TARR50 pKa = 11.84RR51 pKa = 11.84ARR53 pKa = 11.84TTSRR57 pKa = 11.84TTRR60 pKa = 11.84ATHH63 pKa = 6.68RR64 pKa = 11.84ARR66 pKa = 11.84HH67 pKa = 4.56ICRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84NAACLCGRR80 pKa = 11.84AGIGSRR86 pKa = 11.84LL87 pKa = 3.42
Molecular weight: 9.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1360218 |
37 |
2687 |
302.1 |
32.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.292 ± 0.046 | 0.657 ± 0.011 |
5.679 ± 0.03 | 5.715 ± 0.038 |
3.855 ± 0.027 | 8.567 ± 0.034 |
1.918 ± 0.016 | 5.453 ± 0.026 |
3.326 ± 0.031 | 10.158 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.455 ± 0.016 | 2.865 ± 0.025 |
5.14 ± 0.029 | 3.167 ± 0.021 |
6.547 ± 0.041 | 5.399 ± 0.026 |
5.459 ± 0.033 | 7.668 ± 0.029 |
1.344 ± 0.017 | 2.335 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
