
Zavarzinia compransoris
Taxonomy: cellular organisms; Bacteria; Proteobacteria;
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
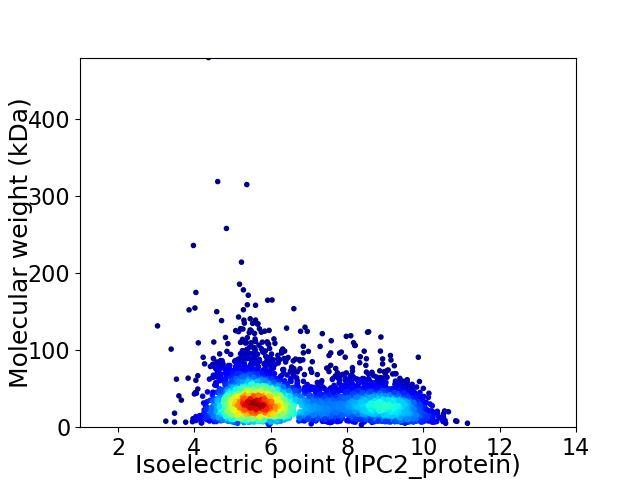
Virtual 2D-PAGE plot for 4405 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A317DT32|A0A317DT32_9PROT 2Fe-2S ferredoxin OS=Zavarzinia compransoris OX=1264899 GN=DKG75_22160 PE=4 SV=1
MM1 pKa = 7.3ATTAAAALSATGFVAAQDD19 pKa = 4.23AYY21 pKa = 9.82PVSGDD26 pKa = 3.12QFGYY30 pKa = 8.17GTRR33 pKa = 11.84AVAEE37 pKa = 4.1TSFNLEE43 pKa = 3.66IFGGADD49 pKa = 3.56EE50 pKa = 5.24NGGTYY55 pKa = 10.47GGAPSITIPLGDD67 pKa = 3.81KK68 pKa = 10.87LGLQVDD74 pKa = 5.01GVAGFTADD82 pKa = 3.19EE83 pKa = 4.48AGFAGGAAQLFYY95 pKa = 10.79RR96 pKa = 11.84DD97 pKa = 4.08PQQGLIGVAAGGYY110 pKa = 7.66YY111 pKa = 9.88VEE113 pKa = 5.82GIKK116 pKa = 10.3QYY118 pKa = 10.77SVAGIAEE125 pKa = 4.43YY126 pKa = 11.21YY127 pKa = 10.26LDD129 pKa = 4.83NITLEE134 pKa = 4.21GLVGYY139 pKa = 7.47QTGDD143 pKa = 3.16VMDD146 pKa = 3.94SVYY149 pKa = 11.03GRR151 pKa = 11.84LGVSIYY157 pKa = 11.14ANPNLRR163 pKa = 11.84LGGGVSYY170 pKa = 11.04SEE172 pKa = 4.05EE173 pKa = 4.09TKK175 pKa = 10.49IGGDD179 pKa = 3.4IQIEE183 pKa = 4.05ALLTDD188 pKa = 3.92VPGLALFATGAFDD201 pKa = 4.03EE202 pKa = 4.9YY203 pKa = 10.97GTMGYY208 pKa = 10.55GGVRR212 pKa = 11.84FYY214 pKa = 11.35FNSGTSLLSTDD225 pKa = 3.49RR226 pKa = 11.84TKK228 pKa = 10.72QAATPSLIDD237 pKa = 2.8MHH239 pKa = 7.11RR240 pKa = 11.84NLGRR244 pKa = 11.84PNFLTNGGAGFGLRR258 pKa = 11.84QISLVGAAKK267 pKa = 10.7NGGDD271 pKa = 4.05DD272 pKa = 4.29FGDD275 pKa = 3.63VTPPPPPPPPPPNPNANCGDD295 pKa = 3.73GLICGVQDD303 pKa = 3.51LVGNLTEE310 pKa = 4.19NTILSPLNDD319 pKa = 5.65LITGLLDD326 pKa = 4.14PNSGALSALTGALEE340 pKa = 4.82DD341 pKa = 4.04LTSSGGGALGAVTDD355 pKa = 4.47LVNGLVNTQNQALEE369 pKa = 4.11PVISGLNSILAQLTGGLTGGLTGGLTGSGATANDD403 pKa = 3.67GLIDD407 pKa = 3.77VVQGVVGNLLDD418 pKa = 3.9GTALEE423 pKa = 4.62GVGDD427 pKa = 4.09LVDD430 pKa = 4.7GLVDD434 pKa = 4.16PNTGALAGLTGALNDD449 pKa = 4.47LTSSDD454 pKa = 4.48AGPLGPVTDD463 pKa = 4.93LVNGLAGANVGALDD477 pKa = 3.75PVLDD481 pKa = 4.39GVHH484 pKa = 7.16DD485 pKa = 4.76LLAGLTGGLAGALPGASSGSTDD507 pKa = 3.01GGLIDD512 pKa = 4.1TVQGTVDD519 pKa = 3.71NLLHH523 pKa = 5.53GTPLEE528 pKa = 4.36VVSDD532 pKa = 3.92LVGGLVDD539 pKa = 4.13PQTGALSALTGQLNNLTAPNAPLGSLTGLVDD570 pKa = 3.99GLVGAQNGALDD581 pKa = 4.07PVLDD585 pKa = 4.17TVNGLLSNNLSSVNLGAAPVVLQNVLPSIPVVGDD619 pKa = 3.65LLSGLVGAIGGIGGG633 pKa = 3.66
MM1 pKa = 7.3ATTAAAALSATGFVAAQDD19 pKa = 4.23AYY21 pKa = 9.82PVSGDD26 pKa = 3.12QFGYY30 pKa = 8.17GTRR33 pKa = 11.84AVAEE37 pKa = 4.1TSFNLEE43 pKa = 3.66IFGGADD49 pKa = 3.56EE50 pKa = 5.24NGGTYY55 pKa = 10.47GGAPSITIPLGDD67 pKa = 3.81KK68 pKa = 10.87LGLQVDD74 pKa = 5.01GVAGFTADD82 pKa = 3.19EE83 pKa = 4.48AGFAGGAAQLFYY95 pKa = 10.79RR96 pKa = 11.84DD97 pKa = 4.08PQQGLIGVAAGGYY110 pKa = 7.66YY111 pKa = 9.88VEE113 pKa = 5.82GIKK116 pKa = 10.3QYY118 pKa = 10.77SVAGIAEE125 pKa = 4.43YY126 pKa = 11.21YY127 pKa = 10.26LDD129 pKa = 4.83NITLEE134 pKa = 4.21GLVGYY139 pKa = 7.47QTGDD143 pKa = 3.16VMDD146 pKa = 3.94SVYY149 pKa = 11.03GRR151 pKa = 11.84LGVSIYY157 pKa = 11.14ANPNLRR163 pKa = 11.84LGGGVSYY170 pKa = 11.04SEE172 pKa = 4.05EE173 pKa = 4.09TKK175 pKa = 10.49IGGDD179 pKa = 3.4IQIEE183 pKa = 4.05ALLTDD188 pKa = 3.92VPGLALFATGAFDD201 pKa = 4.03EE202 pKa = 4.9YY203 pKa = 10.97GTMGYY208 pKa = 10.55GGVRR212 pKa = 11.84FYY214 pKa = 11.35FNSGTSLLSTDD225 pKa = 3.49RR226 pKa = 11.84TKK228 pKa = 10.72QAATPSLIDD237 pKa = 2.8MHH239 pKa = 7.11RR240 pKa = 11.84NLGRR244 pKa = 11.84PNFLTNGGAGFGLRR258 pKa = 11.84QISLVGAAKK267 pKa = 10.7NGGDD271 pKa = 4.05DD272 pKa = 4.29FGDD275 pKa = 3.63VTPPPPPPPPPPNPNANCGDD295 pKa = 3.73GLICGVQDD303 pKa = 3.51LVGNLTEE310 pKa = 4.19NTILSPLNDD319 pKa = 5.65LITGLLDD326 pKa = 4.14PNSGALSALTGALEE340 pKa = 4.82DD341 pKa = 4.04LTSSGGGALGAVTDD355 pKa = 4.47LVNGLVNTQNQALEE369 pKa = 4.11PVISGLNSILAQLTGGLTGGLTGGLTGSGATANDD403 pKa = 3.67GLIDD407 pKa = 3.77VVQGVVGNLLDD418 pKa = 3.9GTALEE423 pKa = 4.62GVGDD427 pKa = 4.09LVDD430 pKa = 4.7GLVDD434 pKa = 4.16PNTGALAGLTGALNDD449 pKa = 4.47LTSSDD454 pKa = 4.48AGPLGPVTDD463 pKa = 4.93LVNGLAGANVGALDD477 pKa = 3.75PVLDD481 pKa = 4.39GVHH484 pKa = 7.16DD485 pKa = 4.76LLAGLTGGLAGALPGASSGSTDD507 pKa = 3.01GGLIDD512 pKa = 4.1TVQGTVDD519 pKa = 3.71NLLHH523 pKa = 5.53GTPLEE528 pKa = 4.36VVSDD532 pKa = 3.92LVGGLVDD539 pKa = 4.13PQTGALSALTGQLNNLTAPNAPLGSLTGLVDD570 pKa = 3.99GLVGAQNGALDD581 pKa = 4.07PVLDD585 pKa = 4.17TVNGLLSNNLSSVNLGAAPVVLQNVLPSIPVVGDD619 pKa = 3.65LLSGLVGAIGGIGGG633 pKa = 3.66
Molecular weight: 62.34 kDa
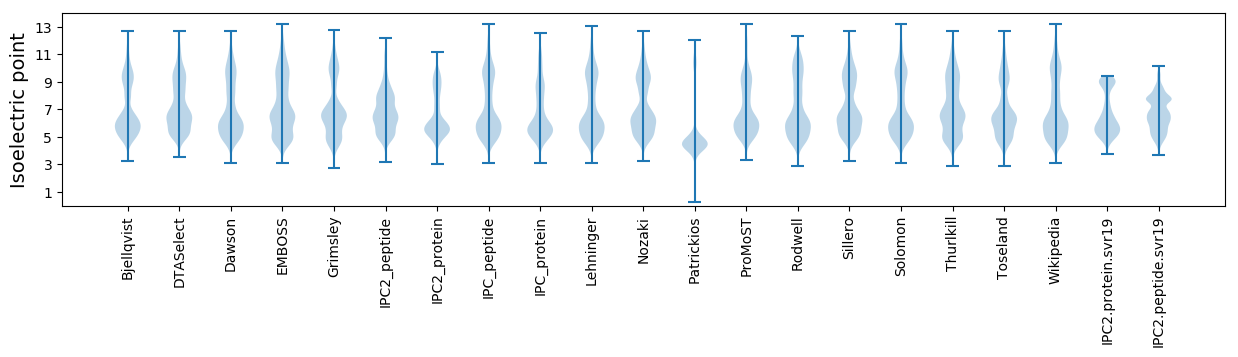
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A317E066|A0A317E066_9PROT ATP-dependent Clp protease ATP-binding subunit ClpA OS=Zavarzinia compransoris OX=1264899 GN=clpA PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.23QPSALVRR12 pKa = 11.84KK13 pKa = 8.86RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.76GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTVGGRR28 pKa = 11.84VVIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.91GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.23QPSALVRR12 pKa = 11.84KK13 pKa = 8.86RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.76GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTVGGRR28 pKa = 11.84VVIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.91GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1447416 |
26 |
4933 |
328.6 |
35.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.236 ± 0.055 | 0.781 ± 0.012 |
5.78 ± 0.028 | 5.271 ± 0.036 |
3.636 ± 0.022 | 9.432 ± 0.051 |
1.964 ± 0.02 | 4.77 ± 0.021 |
2.713 ± 0.028 | 10.697 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.111 ± 0.016 | 2.244 ± 0.024 |
5.59 ± 0.035 | 2.741 ± 0.021 |
7.625 ± 0.042 | 4.404 ± 0.03 |
5.067 ± 0.041 | 7.509 ± 0.037 |
1.288 ± 0.015 | 2.142 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
