
Torque teno midi virus 1 (isolate MD1-073)
Taxonomy: Viruses; Anelloviridae; Gammatorquevirus; Torque teno midi virus 1
Average proteome isoelectric point is 7.16
Get precalculated fractions of proteins
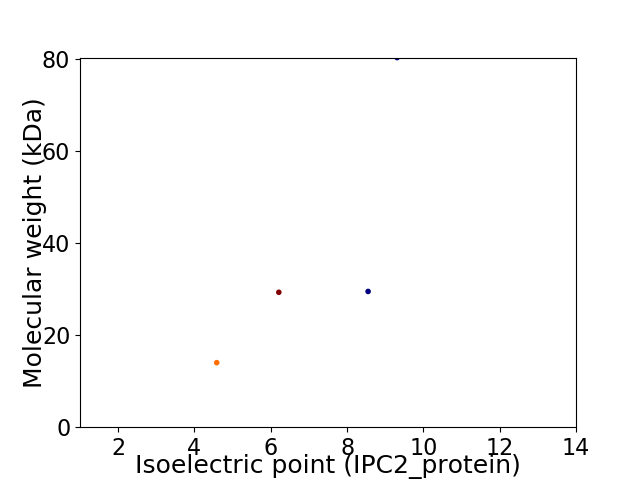
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|A4GZ95-3|ORF3-3_TTVG1 Isoform of A4GZ95 Isoform 3 of Uncharacterized ORF3 protein OS=Torque teno midi virus 1 (isolate MD1-073) OX=766184 GN=ORF3 PE=4 SV=1
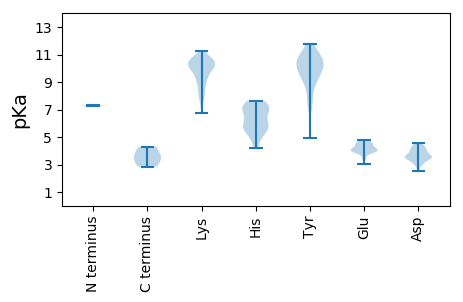
MM1 pKa = 7.26QNLSAKK7 pKa = 10.27DD8 pKa = 3.79FYY10 pKa = 11.44KK11 pKa = 10.12PCRR14 pKa = 11.84YY15 pKa = 9.83NCEE18 pKa = 4.43TKK20 pKa = 10.66NQMWMSGIADD30 pKa = 4.07SHH32 pKa = 7.27DD33 pKa = 3.43SWCDD37 pKa = 2.92CDD39 pKa = 4.31TPFAHH44 pKa = 7.4LLASIFPPGHH54 pKa = 5.8TDD56 pKa = 3.06RR57 pKa = 11.84TRR59 pKa = 11.84TIQEE63 pKa = 3.37ILTRR67 pKa = 11.84DD68 pKa = 3.56FRR70 pKa = 11.84KK71 pKa = 7.97TCLSGGADD79 pKa = 3.35ATNSGMAEE87 pKa = 3.98TIEE90 pKa = 4.29EE91 pKa = 4.18KK92 pKa = 10.79RR93 pKa = 11.84EE94 pKa = 3.83DD95 pKa = 3.6FQKK98 pKa = 10.98EE99 pKa = 3.9EE100 pKa = 4.65KK101 pKa = 10.43EE102 pKa = 4.63DD103 pKa = 3.62FTEE106 pKa = 4.14EE107 pKa = 4.13QNIEE111 pKa = 4.1DD112 pKa = 4.42LLAAVADD119 pKa = 3.93AEE121 pKa = 4.48GRR123 pKa = 3.7
MM1 pKa = 7.26QNLSAKK7 pKa = 10.27DD8 pKa = 3.79FYY10 pKa = 11.44KK11 pKa = 10.12PCRR14 pKa = 11.84YY15 pKa = 9.83NCEE18 pKa = 4.43TKK20 pKa = 10.66NQMWMSGIADD30 pKa = 4.07SHH32 pKa = 7.27DD33 pKa = 3.43SWCDD37 pKa = 2.92CDD39 pKa = 4.31TPFAHH44 pKa = 7.4LLASIFPPGHH54 pKa = 5.8TDD56 pKa = 3.06RR57 pKa = 11.84TRR59 pKa = 11.84TIQEE63 pKa = 3.37ILTRR67 pKa = 11.84DD68 pKa = 3.56FRR70 pKa = 11.84KK71 pKa = 7.97TCLSGGADD79 pKa = 3.35ATNSGMAEE87 pKa = 3.98TIEE90 pKa = 4.29EE91 pKa = 4.18KK92 pKa = 10.79RR93 pKa = 11.84EE94 pKa = 3.83DD95 pKa = 3.6FQKK98 pKa = 10.98EE99 pKa = 3.9EE100 pKa = 4.65KK101 pKa = 10.43EE102 pKa = 4.63DD103 pKa = 3.62FTEE106 pKa = 4.14EE107 pKa = 4.13QNIEE111 pKa = 4.1DD112 pKa = 4.42LLAAVADD119 pKa = 3.93AEE121 pKa = 4.48GRR123 pKa = 3.7
Molecular weight: 14.02 kDa
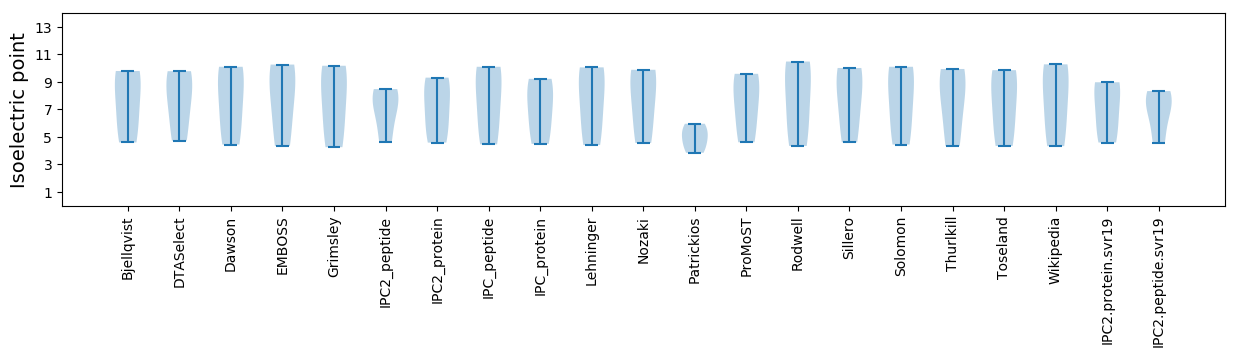
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|A4GZ95-2|ORF3-2_TTVG1 Isoform of A4GZ95 Isoform 2 of Uncharacterized ORF3 protein OS=Torque teno midi virus 1 (isolate MD1-073) OX=766184 GN=ORF3 PE=4 SV=1
MM1 pKa = 7.35PFWWGRR7 pKa = 11.84RR8 pKa = 11.84NKK10 pKa = 9.48FWYY13 pKa = 8.69GRR15 pKa = 11.84NYY17 pKa = 9.7RR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 9.37KK21 pKa = 9.86RR22 pKa = 11.84RR23 pKa = 11.84FPKK26 pKa = 8.58RR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 8.07RR30 pKa = 11.84RR31 pKa = 11.84FYY33 pKa = 10.69RR34 pKa = 11.84RR35 pKa = 11.84TKK37 pKa = 8.83YY38 pKa = 9.96RR39 pKa = 11.84RR40 pKa = 11.84PARR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84KK51 pKa = 8.95VRR53 pKa = 11.84RR54 pKa = 11.84KK55 pKa = 9.75KK56 pKa = 8.97KK57 pKa = 8.65TLIVRR62 pKa = 11.84QWQPDD67 pKa = 4.04SIVLCKK73 pKa = 10.36IKK75 pKa = 10.61GYY77 pKa = 11.1DD78 pKa = 3.55SIIWGAEE85 pKa = 3.73GTQFQCSTHH94 pKa = 5.79EE95 pKa = 4.03MYY97 pKa = 10.47EE98 pKa = 4.13YY99 pKa = 10.48TRR101 pKa = 11.84QKK103 pKa = 10.77YY104 pKa = 9.48PGGGGFGVQLYY115 pKa = 10.08SLEE118 pKa = 3.98YY119 pKa = 10.66LYY121 pKa = 10.71DD122 pKa = 3.33QWKK125 pKa = 9.45LRR127 pKa = 11.84NNIWTKK133 pKa = 9.91TNQLKK138 pKa = 10.4DD139 pKa = 3.13LCRR142 pKa = 11.84YY143 pKa = 7.35LKK145 pKa = 10.71CVMTFYY151 pKa = 10.74RR152 pKa = 11.84HH153 pKa = 4.19QHH155 pKa = 4.83IDD157 pKa = 3.36FVIVYY162 pKa = 7.96EE163 pKa = 4.09RR164 pKa = 11.84QPPFEE169 pKa = 4.57IDD171 pKa = 2.54KK172 pKa = 8.94LTYY175 pKa = 9.08MKK177 pKa = 9.9YY178 pKa = 9.98HH179 pKa = 7.61PYY181 pKa = 9.72MLLQRR186 pKa = 11.84KK187 pKa = 8.47HH188 pKa = 6.78KK189 pKa = 10.31IILPSQTTNPRR200 pKa = 11.84GKK202 pKa = 10.11LKK204 pKa = 10.41KK205 pKa = 9.93KK206 pKa = 9.43KK207 pKa = 8.62TIKK210 pKa = 10.02PPKK213 pKa = 9.77QMLSKK218 pKa = 10.27WFFQQQFAKK227 pKa = 10.74YY228 pKa = 9.53DD229 pKa = 3.81LLLIAAAACSLRR241 pKa = 11.84YY242 pKa = 9.21PRR244 pKa = 11.84IGCCNEE250 pKa = 3.06NRR252 pKa = 11.84MITLYY257 pKa = 10.84CLNTKK262 pKa = 9.85FYY264 pKa = 11.01QDD266 pKa = 4.13TEE268 pKa = 4.02WGTTKK273 pKa = 10.3QAPHH277 pKa = 6.1YY278 pKa = 7.35FKK280 pKa = 10.7PYY282 pKa = 8.64ATINKK287 pKa = 9.79SMIFVSNYY295 pKa = 9.18GGKK298 pKa = 8.07KK299 pKa = 7.44TEE301 pKa = 3.99YY302 pKa = 10.89NIGQWIEE309 pKa = 3.58TDD311 pKa = 3.3IPGEE315 pKa = 4.3GNLARR320 pKa = 11.84YY321 pKa = 8.24YY322 pKa = 10.77RR323 pKa = 11.84SISKK327 pKa = 10.27EE328 pKa = 3.48GGYY331 pKa = 10.15FSPKK335 pKa = 9.55ILQAYY340 pKa = 4.93QTKK343 pKa = 10.1VKK345 pKa = 10.72SVDD348 pKa = 3.67YY349 pKa = 10.41KK350 pKa = 10.64PLPIVLGRR358 pKa = 11.84YY359 pKa = 8.8NPAIDD364 pKa = 4.27DD365 pKa = 4.0GKK367 pKa = 11.26GNKK370 pKa = 9.14IYY372 pKa = 10.58LQTIMNGHH380 pKa = 6.96WGLPQKK386 pKa = 9.76TPDD389 pKa = 3.82YY390 pKa = 10.32IIEE393 pKa = 4.32EE394 pKa = 4.3VPLWLGFWGYY404 pKa = 11.6YY405 pKa = 10.23NYY407 pKa = 10.88LKK409 pKa = 9.02QTRR412 pKa = 11.84TEE414 pKa = 4.57AIFPLHH420 pKa = 5.94MFVVQSKK427 pKa = 9.98YY428 pKa = 10.45IQTQQTEE435 pKa = 4.42TPNNFWAFIDD445 pKa = 3.53NSFIQGKK452 pKa = 8.6NPWDD456 pKa = 3.57SVITYY461 pKa = 9.81SEE463 pKa = 3.89QKK465 pKa = 10.35LWFPTVAWQLKK476 pKa = 7.86TINAICEE483 pKa = 4.05SGPYY487 pKa = 9.18VPKK490 pKa = 10.56LDD492 pKa = 3.54NQTYY496 pKa = 7.58STWEE500 pKa = 3.92LATHH504 pKa = 6.28YY505 pKa = 10.55SFHH508 pKa = 6.68FKK510 pKa = 9.82WGGPQISDD518 pKa = 4.03QPVEE522 pKa = 4.62DD523 pKa = 4.56PGNKK527 pKa = 9.29NKK529 pKa = 10.3YY530 pKa = 9.42DD531 pKa = 3.52VPDD534 pKa = 4.39TIKK537 pKa = 10.76EE538 pKa = 3.83ALQIVNPAKK547 pKa = 10.76NIAATMFHH555 pKa = 6.8DD556 pKa = 3.09WDD558 pKa = 3.76YY559 pKa = 11.74RR560 pKa = 11.84RR561 pKa = 11.84GCITSTAIKK570 pKa = 10.3RR571 pKa = 11.84MQQNLPTDD579 pKa = 3.53SSLEE583 pKa = 3.99SDD585 pKa = 4.22SDD587 pKa = 4.33SEE589 pKa = 4.06PAPKK593 pKa = 9.93KK594 pKa = 10.32KK595 pKa = 10.13RR596 pKa = 11.84LLPVLHH602 pKa = 7.34DD603 pKa = 3.94PQKK606 pKa = 9.96KK607 pKa = 6.76TEE609 pKa = 4.79KK610 pKa = 10.3INQCLLSLCEE620 pKa = 4.27EE621 pKa = 4.61STCQEE626 pKa = 3.95QEE628 pKa = 3.98TEE630 pKa = 4.1EE631 pKa = 4.7NILKK635 pKa = 10.58LIQQQQQQQQKK646 pKa = 9.55LKK648 pKa = 10.67HH649 pKa = 5.85NLLVLIKK656 pKa = 10.17DD657 pKa = 4.01LKK659 pKa = 9.88VKK661 pKa = 10.23QRR663 pKa = 11.84LLQLQTGVLEE673 pKa = 4.3
MM1 pKa = 7.35PFWWGRR7 pKa = 11.84RR8 pKa = 11.84NKK10 pKa = 9.48FWYY13 pKa = 8.69GRR15 pKa = 11.84NYY17 pKa = 9.7RR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 9.37KK21 pKa = 9.86RR22 pKa = 11.84RR23 pKa = 11.84FPKK26 pKa = 8.58RR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 8.07RR30 pKa = 11.84RR31 pKa = 11.84FYY33 pKa = 10.69RR34 pKa = 11.84RR35 pKa = 11.84TKK37 pKa = 8.83YY38 pKa = 9.96RR39 pKa = 11.84RR40 pKa = 11.84PARR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84KK51 pKa = 8.95VRR53 pKa = 11.84RR54 pKa = 11.84KK55 pKa = 9.75KK56 pKa = 8.97KK57 pKa = 8.65TLIVRR62 pKa = 11.84QWQPDD67 pKa = 4.04SIVLCKK73 pKa = 10.36IKK75 pKa = 10.61GYY77 pKa = 11.1DD78 pKa = 3.55SIIWGAEE85 pKa = 3.73GTQFQCSTHH94 pKa = 5.79EE95 pKa = 4.03MYY97 pKa = 10.47EE98 pKa = 4.13YY99 pKa = 10.48TRR101 pKa = 11.84QKK103 pKa = 10.77YY104 pKa = 9.48PGGGGFGVQLYY115 pKa = 10.08SLEE118 pKa = 3.98YY119 pKa = 10.66LYY121 pKa = 10.71DD122 pKa = 3.33QWKK125 pKa = 9.45LRR127 pKa = 11.84NNIWTKK133 pKa = 9.91TNQLKK138 pKa = 10.4DD139 pKa = 3.13LCRR142 pKa = 11.84YY143 pKa = 7.35LKK145 pKa = 10.71CVMTFYY151 pKa = 10.74RR152 pKa = 11.84HH153 pKa = 4.19QHH155 pKa = 4.83IDD157 pKa = 3.36FVIVYY162 pKa = 7.96EE163 pKa = 4.09RR164 pKa = 11.84QPPFEE169 pKa = 4.57IDD171 pKa = 2.54KK172 pKa = 8.94LTYY175 pKa = 9.08MKK177 pKa = 9.9YY178 pKa = 9.98HH179 pKa = 7.61PYY181 pKa = 9.72MLLQRR186 pKa = 11.84KK187 pKa = 8.47HH188 pKa = 6.78KK189 pKa = 10.31IILPSQTTNPRR200 pKa = 11.84GKK202 pKa = 10.11LKK204 pKa = 10.41KK205 pKa = 9.93KK206 pKa = 9.43KK207 pKa = 8.62TIKK210 pKa = 10.02PPKK213 pKa = 9.77QMLSKK218 pKa = 10.27WFFQQQFAKK227 pKa = 10.74YY228 pKa = 9.53DD229 pKa = 3.81LLLIAAAACSLRR241 pKa = 11.84YY242 pKa = 9.21PRR244 pKa = 11.84IGCCNEE250 pKa = 3.06NRR252 pKa = 11.84MITLYY257 pKa = 10.84CLNTKK262 pKa = 9.85FYY264 pKa = 11.01QDD266 pKa = 4.13TEE268 pKa = 4.02WGTTKK273 pKa = 10.3QAPHH277 pKa = 6.1YY278 pKa = 7.35FKK280 pKa = 10.7PYY282 pKa = 8.64ATINKK287 pKa = 9.79SMIFVSNYY295 pKa = 9.18GGKK298 pKa = 8.07KK299 pKa = 7.44TEE301 pKa = 3.99YY302 pKa = 10.89NIGQWIEE309 pKa = 3.58TDD311 pKa = 3.3IPGEE315 pKa = 4.3GNLARR320 pKa = 11.84YY321 pKa = 8.24YY322 pKa = 10.77RR323 pKa = 11.84SISKK327 pKa = 10.27EE328 pKa = 3.48GGYY331 pKa = 10.15FSPKK335 pKa = 9.55ILQAYY340 pKa = 4.93QTKK343 pKa = 10.1VKK345 pKa = 10.72SVDD348 pKa = 3.67YY349 pKa = 10.41KK350 pKa = 10.64PLPIVLGRR358 pKa = 11.84YY359 pKa = 8.8NPAIDD364 pKa = 4.27DD365 pKa = 4.0GKK367 pKa = 11.26GNKK370 pKa = 9.14IYY372 pKa = 10.58LQTIMNGHH380 pKa = 6.96WGLPQKK386 pKa = 9.76TPDD389 pKa = 3.82YY390 pKa = 10.32IIEE393 pKa = 4.32EE394 pKa = 4.3VPLWLGFWGYY404 pKa = 11.6YY405 pKa = 10.23NYY407 pKa = 10.88LKK409 pKa = 9.02QTRR412 pKa = 11.84TEE414 pKa = 4.57AIFPLHH420 pKa = 5.94MFVVQSKK427 pKa = 9.98YY428 pKa = 10.45IQTQQTEE435 pKa = 4.42TPNNFWAFIDD445 pKa = 3.53NSFIQGKK452 pKa = 8.6NPWDD456 pKa = 3.57SVITYY461 pKa = 9.81SEE463 pKa = 3.89QKK465 pKa = 10.35LWFPTVAWQLKK476 pKa = 7.86TINAICEE483 pKa = 4.05SGPYY487 pKa = 9.18VPKK490 pKa = 10.56LDD492 pKa = 3.54NQTYY496 pKa = 7.58STWEE500 pKa = 3.92LATHH504 pKa = 6.28YY505 pKa = 10.55SFHH508 pKa = 6.68FKK510 pKa = 9.82WGGPQISDD518 pKa = 4.03QPVEE522 pKa = 4.62DD523 pKa = 4.56PGNKK527 pKa = 9.29NKK529 pKa = 10.3YY530 pKa = 9.42DD531 pKa = 3.52VPDD534 pKa = 4.39TIKK537 pKa = 10.76EE538 pKa = 3.83ALQIVNPAKK547 pKa = 10.76NIAATMFHH555 pKa = 6.8DD556 pKa = 3.09WDD558 pKa = 3.76YY559 pKa = 11.74RR560 pKa = 11.84RR561 pKa = 11.84GCITSTAIKK570 pKa = 10.3RR571 pKa = 11.84MQQNLPTDD579 pKa = 3.53SSLEE583 pKa = 3.99SDD585 pKa = 4.22SDD587 pKa = 4.33SEE589 pKa = 4.06PAPKK593 pKa = 9.93KK594 pKa = 10.32KK595 pKa = 10.13RR596 pKa = 11.84LLPVLHH602 pKa = 7.34DD603 pKa = 3.94PQKK606 pKa = 9.96KK607 pKa = 6.76TEE609 pKa = 4.79KK610 pKa = 10.3INQCLLSLCEE620 pKa = 4.27EE621 pKa = 4.61STCQEE626 pKa = 3.95QEE628 pKa = 3.98TEE630 pKa = 4.1EE631 pKa = 4.7NILKK635 pKa = 10.58LIQQQQQQQQKK646 pKa = 9.55LKK648 pKa = 10.67HH649 pKa = 5.85NLLVLIKK656 pKa = 10.17DD657 pKa = 4.01LKK659 pKa = 9.88VKK661 pKa = 10.23QRR663 pKa = 11.84LLQLQTGVLEE673 pKa = 4.3
Molecular weight: 80.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1316 |
123 |
673 |
329.0 |
38.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.395 ± 1.477 | 2.356 ± 0.423 |
5.091 ± 0.988 | 6.155 ± 1.251 |
3.951 ± 0.496 | 4.863 ± 0.489 |
2.356 ± 0.322 | 5.243 ± 0.943 |
9.043 ± 0.874 | 7.447 ± 0.545 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.204 ± 0.315 | 4.559 ± 0.084 |
5.319 ± 0.917 | 6.687 ± 0.828 |
6.383 ± 0.255 | 6.231 ± 1.693 |
8.131 ± 1.143 | 2.356 ± 0.82 |
2.052 ± 0.599 | 4.179 ± 1.473 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
