
Bat coronavirus 512/2005 (BtCoV) (BtCoV/512/2005)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Pedacovirus
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
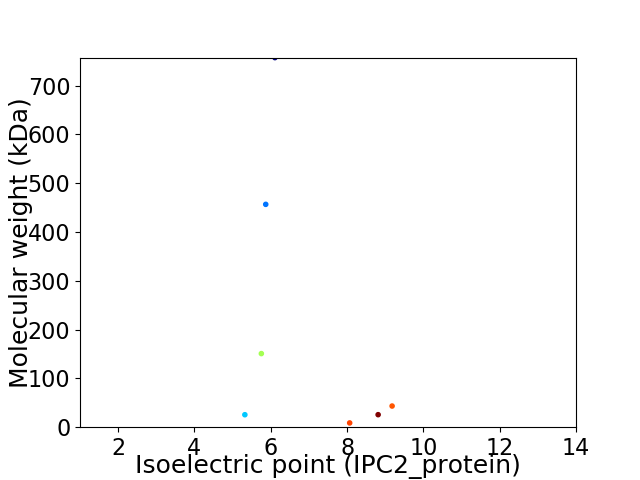
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
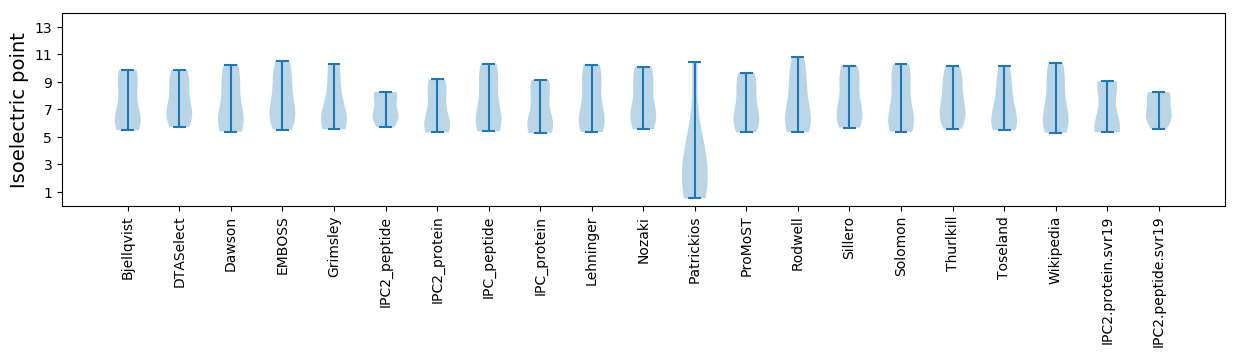
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q0Q466|SPIKE_BC512 Spike glycoprotein OS=Bat coronavirus 512/2005 OX=693999 GN=S PE=3 SV=1
MM1 pKa = 7.48FLGLFQYY8 pKa = 10.55TIDD11 pKa = 3.56TAVEE15 pKa = 4.13HH16 pKa = 5.72TVEE19 pKa = 4.19HH20 pKa = 6.71ANLSQEE26 pKa = 4.13EE27 pKa = 4.01ALMLEE32 pKa = 4.53EE33 pKa = 5.03NIVPLRR39 pKa = 11.84QATHH43 pKa = 3.98VTGFLLTSVFVYY55 pKa = 8.62FFALFKK61 pKa = 10.8ASSYY65 pKa = 10.53KK66 pKa = 10.47RR67 pKa = 11.84NLLLFLARR75 pKa = 11.84LLALLIYY82 pKa = 10.55APILIFCGAYY92 pKa = 9.67LDD94 pKa = 4.08AFIVVATLTSRR105 pKa = 11.84LLFLTYY111 pKa = 10.13YY112 pKa = 8.13SWRR115 pKa = 11.84YY116 pKa = 7.08KK117 pKa = 9.3TYY119 pKa = 10.84KK120 pKa = 10.02FLIYY124 pKa = 10.5NSSTLMFLHH133 pKa = 6.49GHH135 pKa = 6.01ANYY138 pKa = 10.72YY139 pKa = 9.83NGRR142 pKa = 11.84PYY144 pKa = 11.59VMLEE148 pKa = 4.08GGSHH152 pKa = 5.67YY153 pKa = 10.09VTLGTDD159 pKa = 3.41IVPFVSRR166 pKa = 11.84SNLYY170 pKa = 9.95LAIRR174 pKa = 11.84GSAEE178 pKa = 3.87SDD180 pKa = 2.98IQLLRR185 pKa = 11.84TVEE188 pKa = 4.15LLDD191 pKa = 3.99GNYY194 pKa = 10.59LYY196 pKa = 10.24IFSSCQVVGVTNSGFEE212 pKa = 4.14EE213 pKa = 4.15IQLDD217 pKa = 4.2EE218 pKa = 4.28YY219 pKa = 11.22ATISEE224 pKa = 4.39
MM1 pKa = 7.48FLGLFQYY8 pKa = 10.55TIDD11 pKa = 3.56TAVEE15 pKa = 4.13HH16 pKa = 5.72TVEE19 pKa = 4.19HH20 pKa = 6.71ANLSQEE26 pKa = 4.13EE27 pKa = 4.01ALMLEE32 pKa = 4.53EE33 pKa = 5.03NIVPLRR39 pKa = 11.84QATHH43 pKa = 3.98VTGFLLTSVFVYY55 pKa = 8.62FFALFKK61 pKa = 10.8ASSYY65 pKa = 10.53KK66 pKa = 10.47RR67 pKa = 11.84NLLLFLARR75 pKa = 11.84LLALLIYY82 pKa = 10.55APILIFCGAYY92 pKa = 9.67LDD94 pKa = 4.08AFIVVATLTSRR105 pKa = 11.84LLFLTYY111 pKa = 10.13YY112 pKa = 8.13SWRR115 pKa = 11.84YY116 pKa = 7.08KK117 pKa = 9.3TYY119 pKa = 10.84KK120 pKa = 10.02FLIYY124 pKa = 10.5NSSTLMFLHH133 pKa = 6.49GHH135 pKa = 6.01ANYY138 pKa = 10.72YY139 pKa = 9.83NGRR142 pKa = 11.84PYY144 pKa = 11.59VMLEE148 pKa = 4.08GGSHH152 pKa = 5.67YY153 pKa = 10.09VTLGTDD159 pKa = 3.41IVPFVSRR166 pKa = 11.84SNLYY170 pKa = 9.95LAIRR174 pKa = 11.84GSAEE178 pKa = 3.87SDD180 pKa = 2.98IQLLRR185 pKa = 11.84TVEE188 pKa = 4.15LLDD191 pKa = 3.99GNYY194 pKa = 10.59LYY196 pKa = 10.24IFSSCQVVGVTNSGFEE212 pKa = 4.14EE213 pKa = 4.15IQLDD217 pKa = 4.2EE218 pKa = 4.28YY219 pKa = 11.22ATISEE224 pKa = 4.39
Molecular weight: 25.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q0Q463|VME1_BC512 Membrane protein OS=Bat coronavirus 512/2005 OX=693999 GN=M PE=3 SV=1
MM1 pKa = 7.67ASVKK5 pKa = 9.27FQPRR9 pKa = 11.84GRR11 pKa = 11.84SKK13 pKa = 11.13GRR15 pKa = 11.84VPLSLFAPLRR25 pKa = 11.84VTDD28 pKa = 4.44EE29 pKa = 4.06KK30 pKa = 10.97PLYY33 pKa = 10.13KK34 pKa = 10.28VLPNNAVPQGMGGKK48 pKa = 9.05DD49 pKa = 3.18QQIGYY54 pKa = 8.72WVEE57 pKa = 3.76QQRR60 pKa = 11.84WRR62 pKa = 11.84MRR64 pKa = 11.84RR65 pKa = 11.84GDD67 pKa = 4.01RR68 pKa = 11.84VDD70 pKa = 5.53LPSNWHH76 pKa = 6.43FYY78 pKa = 11.01FLGTGPHH85 pKa = 6.48SDD87 pKa = 3.13LPFRR91 pKa = 11.84KK92 pKa = 8.78RR93 pKa = 11.84TDD95 pKa = 3.05GVFWVAIDD103 pKa = 3.88GAKK106 pKa = 8.22TQPTGLGVRR115 pKa = 11.84KK116 pKa = 10.04SSEE119 pKa = 3.92KK120 pKa = 10.41PLVPKK125 pKa = 10.46FKK127 pKa = 11.02NKK129 pKa = 10.07LPNNVEE135 pKa = 3.87IVEE138 pKa = 4.27PTTPNNSRR146 pKa = 11.84ANSRR150 pKa = 11.84SRR152 pKa = 11.84SRR154 pKa = 11.84GGQSNSRR161 pKa = 11.84GNSQNRR167 pKa = 11.84GDD169 pKa = 3.61KK170 pKa = 10.65SRR172 pKa = 11.84NQSRR176 pKa = 11.84NRR178 pKa = 11.84SQSNDD183 pKa = 3.28RR184 pKa = 11.84GSDD187 pKa = 3.4SRR189 pKa = 11.84DD190 pKa = 3.43DD191 pKa = 3.64LVAAVKK197 pKa = 10.31KK198 pKa = 10.49ALEE201 pKa = 4.24DD202 pKa = 3.91LGVGAAKK209 pKa = 10.36PKK211 pKa = 10.86GKK213 pKa = 7.85TQSGKK218 pKa = 8.26NTPKK222 pKa = 10.31NKK224 pKa = 9.62SRR226 pKa = 11.84SGSVQRR232 pKa = 11.84AEE234 pKa = 5.3AKK236 pKa = 10.11DD237 pKa = 3.43KK238 pKa = 10.96PEE240 pKa = 3.49WRR242 pKa = 11.84RR243 pKa = 11.84TPSGDD248 pKa = 3.18EE249 pKa = 3.96SVEE252 pKa = 4.08VCFGPRR258 pKa = 11.84GGTRR262 pKa = 11.84NFGSSEE268 pKa = 3.91FVAKK272 pKa = 10.21GVNAPGYY279 pKa = 8.95AQAASLVPGAAALLFGGNVATKK301 pKa = 10.21EE302 pKa = 4.0MADD305 pKa = 3.62GVEE308 pKa = 3.54ITYY311 pKa = 8.85TYY313 pKa = 11.13KK314 pKa = 10.29MLVPKK319 pKa = 10.12DD320 pKa = 4.0DD321 pKa = 4.07KK322 pKa = 11.38NLEE325 pKa = 3.86IFLAQVDD332 pKa = 4.47AYY334 pKa = 10.16KK335 pKa = 10.93LGDD338 pKa = 3.84PKK340 pKa = 10.54PQRR343 pKa = 11.84KK344 pKa = 8.55VKK346 pKa = 10.45RR347 pKa = 11.84SRR349 pKa = 11.84TPTPKK354 pKa = 10.02PATEE358 pKa = 3.82PVYY361 pKa = 11.2DD362 pKa = 5.21DD363 pKa = 3.74VAADD367 pKa = 3.41PTYY370 pKa = 11.66ANLEE374 pKa = 3.87WDD376 pKa = 3.74TTVEE380 pKa = 4.85DD381 pKa = 4.1GVEE384 pKa = 4.22MINEE388 pKa = 4.04VFDD391 pKa = 3.88TQNN394 pKa = 2.67
MM1 pKa = 7.67ASVKK5 pKa = 9.27FQPRR9 pKa = 11.84GRR11 pKa = 11.84SKK13 pKa = 11.13GRR15 pKa = 11.84VPLSLFAPLRR25 pKa = 11.84VTDD28 pKa = 4.44EE29 pKa = 4.06KK30 pKa = 10.97PLYY33 pKa = 10.13KK34 pKa = 10.28VLPNNAVPQGMGGKK48 pKa = 9.05DD49 pKa = 3.18QQIGYY54 pKa = 8.72WVEE57 pKa = 3.76QQRR60 pKa = 11.84WRR62 pKa = 11.84MRR64 pKa = 11.84RR65 pKa = 11.84GDD67 pKa = 4.01RR68 pKa = 11.84VDD70 pKa = 5.53LPSNWHH76 pKa = 6.43FYY78 pKa = 11.01FLGTGPHH85 pKa = 6.48SDD87 pKa = 3.13LPFRR91 pKa = 11.84KK92 pKa = 8.78RR93 pKa = 11.84TDD95 pKa = 3.05GVFWVAIDD103 pKa = 3.88GAKK106 pKa = 8.22TQPTGLGVRR115 pKa = 11.84KK116 pKa = 10.04SSEE119 pKa = 3.92KK120 pKa = 10.41PLVPKK125 pKa = 10.46FKK127 pKa = 11.02NKK129 pKa = 10.07LPNNVEE135 pKa = 3.87IVEE138 pKa = 4.27PTTPNNSRR146 pKa = 11.84ANSRR150 pKa = 11.84SRR152 pKa = 11.84SRR154 pKa = 11.84GGQSNSRR161 pKa = 11.84GNSQNRR167 pKa = 11.84GDD169 pKa = 3.61KK170 pKa = 10.65SRR172 pKa = 11.84NQSRR176 pKa = 11.84NRR178 pKa = 11.84SQSNDD183 pKa = 3.28RR184 pKa = 11.84GSDD187 pKa = 3.4SRR189 pKa = 11.84DD190 pKa = 3.43DD191 pKa = 3.64LVAAVKK197 pKa = 10.31KK198 pKa = 10.49ALEE201 pKa = 4.24DD202 pKa = 3.91LGVGAAKK209 pKa = 10.36PKK211 pKa = 10.86GKK213 pKa = 7.85TQSGKK218 pKa = 8.26NTPKK222 pKa = 10.31NKK224 pKa = 9.62SRR226 pKa = 11.84SGSVQRR232 pKa = 11.84AEE234 pKa = 5.3AKK236 pKa = 10.11DD237 pKa = 3.43KK238 pKa = 10.96PEE240 pKa = 3.49WRR242 pKa = 11.84RR243 pKa = 11.84TPSGDD248 pKa = 3.18EE249 pKa = 3.96SVEE252 pKa = 4.08VCFGPRR258 pKa = 11.84GGTRR262 pKa = 11.84NFGSSEE268 pKa = 3.91FVAKK272 pKa = 10.21GVNAPGYY279 pKa = 8.95AQAASLVPGAAALLFGGNVATKK301 pKa = 10.21EE302 pKa = 4.0MADD305 pKa = 3.62GVEE308 pKa = 3.54ITYY311 pKa = 8.85TYY313 pKa = 11.13KK314 pKa = 10.29MLVPKK319 pKa = 10.12DD320 pKa = 4.0DD321 pKa = 4.07KK322 pKa = 11.38NLEE325 pKa = 3.86IFLAQVDD332 pKa = 4.47AYY334 pKa = 10.16KK335 pKa = 10.93LGDD338 pKa = 3.84PKK340 pKa = 10.54PQRR343 pKa = 11.84KK344 pKa = 8.55VKK346 pKa = 10.45RR347 pKa = 11.84SRR349 pKa = 11.84TPTPKK354 pKa = 10.02PATEE358 pKa = 3.82PVYY361 pKa = 11.2DD362 pKa = 5.21DD363 pKa = 3.74VAADD367 pKa = 3.41PTYY370 pKa = 11.66ANLEE374 pKa = 3.87WDD376 pKa = 3.74TTVEE380 pKa = 4.85DD381 pKa = 4.1GVEE384 pKa = 4.22MINEE388 pKa = 4.04VFDD391 pKa = 3.88TQNN394 pKa = 2.67
Molecular weight: 43.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13213 |
76 |
6793 |
1887.6 |
209.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.259 ± 0.191 | 3.277 ± 0.401 |
5.752 ± 0.545 | 3.837 ± 0.155 |
5.601 ± 0.157 | 6.751 ± 0.229 |
1.862 ± 0.157 | 4.859 ± 0.185 |
5.775 ± 0.595 | 9.362 ± 0.644 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.937 ± 0.109 | 5.661 ± 0.359 |
3.64 ± 0.238 | 3.035 ± 0.307 |
3.459 ± 0.25 | 7.144 ± 0.169 |
6.229 ± 0.42 | 10.073 ± 0.215 |
1.143 ± 0.207 | 4.344 ± 0.312 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
