
Microbacterium sp. TS-1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium; unclassified Microbacterium
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
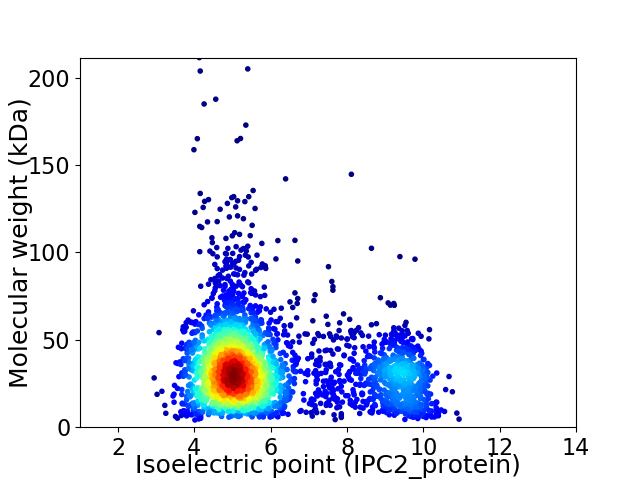
Virtual 2D-PAGE plot for 3155 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U2YQA0|U2YQA0_9MICO ABC transporter ATP-binding protein OS=Microbacterium sp. TS-1 OX=1344956 GN=MTS1_03148 PE=4 SV=1
MM1 pKa = 7.88PSLTSPRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84TLLGVTVALAAALALSSCAAGTSSSEE36 pKa = 4.4DD37 pKa = 3.48GEE39 pKa = 4.31QTLVIGSQMASFPSLDD55 pKa = 3.17TGAITTAGYY64 pKa = 9.71EE65 pKa = 4.25GQRR68 pKa = 11.84LIGNLVYY75 pKa = 10.75EE76 pKa = 4.84GLTKK80 pKa = 10.05RR81 pKa = 11.84DD82 pKa = 3.39VSDD85 pKa = 4.2PNAPAGVAPALASSWEE101 pKa = 4.01VADD104 pKa = 5.68DD105 pKa = 3.91GLSYY109 pKa = 10.92TFTLQDD115 pKa = 2.9GVTFHH120 pKa = 7.87DD121 pKa = 4.3GTDD124 pKa = 3.11WDD126 pKa = 4.45ADD128 pKa = 3.57ALIYY132 pKa = 10.51NINRR136 pKa = 11.84YY137 pKa = 7.95MDD139 pKa = 3.79EE140 pKa = 4.19TDD142 pKa = 3.51PNYY145 pKa = 11.14DD146 pKa = 3.4PVLITYY152 pKa = 7.41YY153 pKa = 10.23TVLNFVDD160 pKa = 4.46SVEE163 pKa = 4.6KK164 pKa = 10.78VDD166 pKa = 5.38DD167 pKa = 3.64MTVTFTLNQPFAFFLADD184 pKa = 4.28LYY186 pKa = 11.36NVYY189 pKa = 9.94FASPTSLDD197 pKa = 3.3EE198 pKa = 5.76DD199 pKa = 4.25GAAGQATNPVGTGPFVFDD217 pKa = 4.69EE218 pKa = 4.32LQQNQSISFVRR229 pKa = 11.84NDD231 pKa = 4.13DD232 pKa = 3.3YY233 pKa = 11.14WGEE236 pKa = 4.2VPQLDD241 pKa = 4.03RR242 pKa = 11.84LTVEE246 pKa = 5.58LIPDD250 pKa = 4.17PSARR254 pKa = 11.84TAALRR259 pKa = 11.84SGRR262 pKa = 11.84VDD264 pKa = 3.34WIEE267 pKa = 3.77AAQPDD272 pKa = 4.54DD273 pKa = 4.01LASLEE278 pKa = 4.33SAGFVATGNAFDD290 pKa = 5.37WEE292 pKa = 4.5WSWQLMTGEE301 pKa = 4.96APFDD305 pKa = 4.89DD306 pKa = 3.36IRR308 pKa = 11.84VRR310 pKa = 11.84QAMNYY315 pKa = 10.07AIDD318 pKa = 3.87RR319 pKa = 11.84EE320 pKa = 4.47AIADD324 pKa = 3.99DD325 pKa = 4.55LLHH328 pKa = 5.92GTAVPAHH335 pKa = 5.78QMFASASLYY344 pKa = 8.79YY345 pKa = 10.52TEE347 pKa = 6.57DD348 pKa = 4.25DD349 pKa = 4.74DD350 pKa = 6.72LYY352 pKa = 11.32DD353 pKa = 4.31YY354 pKa = 11.39DD355 pKa = 4.13PEE357 pKa = 4.2KK358 pKa = 11.11AKK360 pKa = 10.96ALLAEE365 pKa = 4.61AGYY368 pKa = 9.9PDD370 pKa = 4.42GLDD373 pKa = 3.18VTVGYY378 pKa = 8.45ITAGSGAMQAKK389 pKa = 9.49SMNEE393 pKa = 3.52ALQTQLAAVGINVSLEE409 pKa = 3.98PVDD412 pKa = 4.11FSGMYY417 pKa = 10.38AKK419 pKa = 10.53LGEE422 pKa = 4.88GDD424 pKa = 4.24TGWQAANQAFSLEE437 pKa = 4.6QPSSWNGFWFGCDD450 pKa = 3.0GNFFGYY456 pKa = 10.46CNPEE460 pKa = 3.81AEE462 pKa = 4.32ALYY465 pKa = 10.44DD466 pKa = 3.68QALTTVDD473 pKa = 3.66DD474 pKa = 4.3EE475 pKa = 4.39EE476 pKa = 4.49RR477 pKa = 11.84AALLTEE483 pKa = 4.23VGHH486 pKa = 7.13LVTEE490 pKa = 4.48DD491 pKa = 3.64AAWLFVVNDD500 pKa = 3.84TAPRR504 pKa = 11.84AMSSAVTGYY513 pKa = 7.65EE514 pKa = 3.99QPKK517 pKa = 9.7SWWIDD522 pKa = 3.25FTGISMGDD530 pKa = 3.04
MM1 pKa = 7.88PSLTSPRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84TLLGVTVALAAALALSSCAAGTSSSEE36 pKa = 4.4DD37 pKa = 3.48GEE39 pKa = 4.31QTLVIGSQMASFPSLDD55 pKa = 3.17TGAITTAGYY64 pKa = 9.71EE65 pKa = 4.25GQRR68 pKa = 11.84LIGNLVYY75 pKa = 10.75EE76 pKa = 4.84GLTKK80 pKa = 10.05RR81 pKa = 11.84DD82 pKa = 3.39VSDD85 pKa = 4.2PNAPAGVAPALASSWEE101 pKa = 4.01VADD104 pKa = 5.68DD105 pKa = 3.91GLSYY109 pKa = 10.92TFTLQDD115 pKa = 2.9GVTFHH120 pKa = 7.87DD121 pKa = 4.3GTDD124 pKa = 3.11WDD126 pKa = 4.45ADD128 pKa = 3.57ALIYY132 pKa = 10.51NINRR136 pKa = 11.84YY137 pKa = 7.95MDD139 pKa = 3.79EE140 pKa = 4.19TDD142 pKa = 3.51PNYY145 pKa = 11.14DD146 pKa = 3.4PVLITYY152 pKa = 7.41YY153 pKa = 10.23TVLNFVDD160 pKa = 4.46SVEE163 pKa = 4.6KK164 pKa = 10.78VDD166 pKa = 5.38DD167 pKa = 3.64MTVTFTLNQPFAFFLADD184 pKa = 4.28LYY186 pKa = 11.36NVYY189 pKa = 9.94FASPTSLDD197 pKa = 3.3EE198 pKa = 5.76DD199 pKa = 4.25GAAGQATNPVGTGPFVFDD217 pKa = 4.69EE218 pKa = 4.32LQQNQSISFVRR229 pKa = 11.84NDD231 pKa = 4.13DD232 pKa = 3.3YY233 pKa = 11.14WGEE236 pKa = 4.2VPQLDD241 pKa = 4.03RR242 pKa = 11.84LTVEE246 pKa = 5.58LIPDD250 pKa = 4.17PSARR254 pKa = 11.84TAALRR259 pKa = 11.84SGRR262 pKa = 11.84VDD264 pKa = 3.34WIEE267 pKa = 3.77AAQPDD272 pKa = 4.54DD273 pKa = 4.01LASLEE278 pKa = 4.33SAGFVATGNAFDD290 pKa = 5.37WEE292 pKa = 4.5WSWQLMTGEE301 pKa = 4.96APFDD305 pKa = 4.89DD306 pKa = 3.36IRR308 pKa = 11.84VRR310 pKa = 11.84QAMNYY315 pKa = 10.07AIDD318 pKa = 3.87RR319 pKa = 11.84EE320 pKa = 4.47AIADD324 pKa = 3.99DD325 pKa = 4.55LLHH328 pKa = 5.92GTAVPAHH335 pKa = 5.78QMFASASLYY344 pKa = 8.79YY345 pKa = 10.52TEE347 pKa = 6.57DD348 pKa = 4.25DD349 pKa = 4.74DD350 pKa = 6.72LYY352 pKa = 11.32DD353 pKa = 4.31YY354 pKa = 11.39DD355 pKa = 4.13PEE357 pKa = 4.2KK358 pKa = 11.11AKK360 pKa = 10.96ALLAEE365 pKa = 4.61AGYY368 pKa = 9.9PDD370 pKa = 4.42GLDD373 pKa = 3.18VTVGYY378 pKa = 8.45ITAGSGAMQAKK389 pKa = 9.49SMNEE393 pKa = 3.52ALQTQLAAVGINVSLEE409 pKa = 3.98PVDD412 pKa = 4.11FSGMYY417 pKa = 10.38AKK419 pKa = 10.53LGEE422 pKa = 4.88GDD424 pKa = 4.24TGWQAANQAFSLEE437 pKa = 4.6QPSSWNGFWFGCDD450 pKa = 3.0GNFFGYY456 pKa = 10.46CNPEE460 pKa = 3.81AEE462 pKa = 4.32ALYY465 pKa = 10.44DD466 pKa = 3.68QALTTVDD473 pKa = 3.66DD474 pKa = 4.3EE475 pKa = 4.39EE476 pKa = 4.49RR477 pKa = 11.84AALLTEE483 pKa = 4.23VGHH486 pKa = 7.13LVTEE490 pKa = 4.48DD491 pKa = 3.64AAWLFVVNDD500 pKa = 3.84TAPRR504 pKa = 11.84AMSSAVTGYY513 pKa = 7.65EE514 pKa = 3.99QPKK517 pKa = 9.7SWWIDD522 pKa = 3.25FTGISMGDD530 pKa = 3.04
Molecular weight: 57.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U2XNR2|U2XNR2_9MICO Dipeptide/oligopeptide/nickel ABC transporter permease protein OS=Microbacterium sp. TS-1 OX=1344956 GN=MTS1_02072 PE=4 SV=1
MM1 pKa = 7.7PARR4 pKa = 11.84AVLYY8 pKa = 9.86PRR10 pKa = 11.84VPGLKK15 pKa = 9.65RR16 pKa = 11.84SGDD19 pKa = 3.41RR20 pKa = 11.84RR21 pKa = 11.84SGVIPMSKK29 pKa = 8.84RR30 pKa = 11.84TFQPNNRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84AKK41 pKa = 10.04KK42 pKa = 8.86HH43 pKa = 4.36GFRR46 pKa = 11.84ARR48 pKa = 11.84MRR50 pKa = 11.84TRR52 pKa = 11.84AGRR55 pKa = 11.84AILSARR61 pKa = 11.84RR62 pKa = 11.84GKK64 pKa = 10.71GRR66 pKa = 11.84TEE68 pKa = 4.13LSAA71 pKa = 4.86
MM1 pKa = 7.7PARR4 pKa = 11.84AVLYY8 pKa = 9.86PRR10 pKa = 11.84VPGLKK15 pKa = 9.65RR16 pKa = 11.84SGDD19 pKa = 3.41RR20 pKa = 11.84RR21 pKa = 11.84SGVIPMSKK29 pKa = 8.84RR30 pKa = 11.84TFQPNNRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84AKK41 pKa = 10.04KK42 pKa = 8.86HH43 pKa = 4.36GFRR46 pKa = 11.84ARR48 pKa = 11.84MRR50 pKa = 11.84TRR52 pKa = 11.84AGRR55 pKa = 11.84AILSARR61 pKa = 11.84RR62 pKa = 11.84GKK64 pKa = 10.71GRR66 pKa = 11.84TEE68 pKa = 4.13LSAA71 pKa = 4.86
Molecular weight: 8.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1046449 |
40 |
2022 |
331.7 |
35.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.084 ± 0.061 | 0.466 ± 0.009 |
6.471 ± 0.044 | 5.511 ± 0.042 |
3.105 ± 0.027 | 9.003 ± 0.038 |
1.896 ± 0.021 | 4.49 ± 0.032 |
1.609 ± 0.033 | 9.961 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.736 ± 0.018 | 1.81 ± 0.022 |
5.492 ± 0.033 | 2.589 ± 0.024 |
7.684 ± 0.051 | 5.569 ± 0.029 |
5.944 ± 0.036 | 9.138 ± 0.043 |
1.522 ± 0.022 | 1.919 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
