
Fathead minnow nidovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Tornidovirineae; Tobaniviridae; Piscanivirinae; Bafinivirus; Pimfabavirus; Fathead minnow nidovirus 1
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
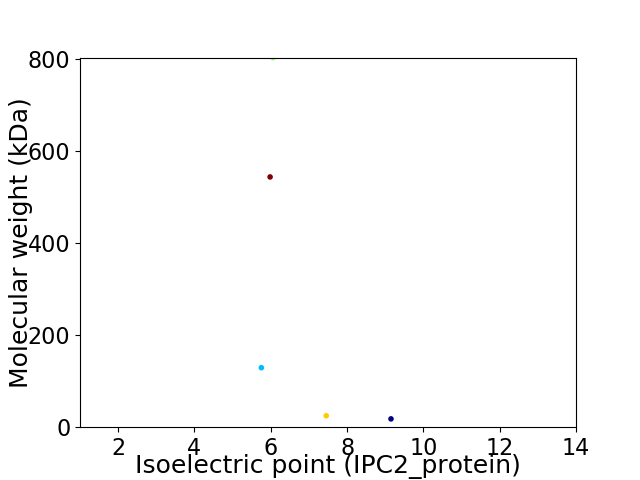
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I6LMN2|I6LMN2_9NIDO Membrane protein OS=Fathead minnow nidovirus OX=889873 GN=M PE=4 SV=1
MM1 pKa = 7.31MLLILPLILLCSSTHH16 pKa = 6.47AYY18 pKa = 9.16RR19 pKa = 11.84VQFVAPVPLNWINCYY34 pKa = 7.76TTDD37 pKa = 3.64CVSWQACPTTVAALVCVRR55 pKa = 11.84FTAVAPSFYY64 pKa = 10.8LSANNTLIGYY74 pKa = 8.4NADD77 pKa = 3.46PNTFYY82 pKa = 11.51DD83 pKa = 5.49LITMVQQSTYY93 pKa = 10.88SFSQAQTDD101 pKa = 3.92TPPTKK106 pKa = 10.18QCNGPSTILSGQCLTPADD124 pKa = 3.95SCLDD128 pKa = 3.86PLCPTTSFFEE138 pKa = 4.33TGSLDD143 pKa = 4.07SLTLDD148 pKa = 3.84TQCVASYY155 pKa = 10.32KK156 pKa = 10.64GYY158 pKa = 9.81GSVYY162 pKa = 9.03GTPCEE167 pKa = 4.02PFAPQLSPYY176 pKa = 7.91FTKK179 pKa = 10.68NVPNTPGFTDD189 pKa = 3.36YY190 pKa = 11.21GVNRR194 pKa = 11.84QSPKK198 pKa = 9.9LGCYY202 pKa = 7.25YY203 pKa = 10.21EE204 pKa = 4.71KK205 pKa = 9.75WAKK208 pKa = 9.84YY209 pKa = 9.42RR210 pKa = 11.84PKK212 pKa = 10.34PYY214 pKa = 9.56TATIDD219 pKa = 3.68SLAGCDD225 pKa = 2.8IVFFFVNTLSDD236 pKa = 3.11RR237 pKa = 11.84SGNMGVFEE245 pKa = 4.99RR246 pKa = 11.84EE247 pKa = 3.64EE248 pKa = 4.63GDD250 pKa = 3.32WNTIAAIKK258 pKa = 9.64TKK260 pKa = 10.47FPTTQVFLTFGGWTSDD276 pKa = 3.44NKK278 pKa = 10.52IMSAAMSNSTILNDD292 pKa = 3.06INTLSSRR299 pKa = 11.84LGVSVDD305 pKa = 4.06FDD307 pKa = 3.78IEE309 pKa = 4.51FPGSSNGNAAVFPYY323 pKa = 10.67DD324 pKa = 3.42EE325 pKa = 4.64ALITSFMTDD334 pKa = 1.69ICNFQHH340 pKa = 6.82SAGRR344 pKa = 11.84QCAWGGLSVGSHH356 pKa = 5.3WHH358 pKa = 5.49PTLINKK364 pKa = 9.75LSTIAGVDD372 pKa = 3.23YY373 pKa = 10.58FPIFGYY379 pKa = 10.34DD380 pKa = 3.3LHH382 pKa = 7.79GSWAPIPRR390 pKa = 11.84QQSALVNYY398 pKa = 10.27SPDD401 pKa = 3.45PTVSGGGPLNTYY413 pKa = 9.96SLITSVDD420 pKa = 3.2NFLTIVPASKK430 pKa = 10.71LILGLPMYY438 pKa = 10.83ARR440 pKa = 11.84GYY442 pKa = 10.78LVDD445 pKa = 4.01SSNSVLGSFPLTGPMLGPGEE465 pKa = 4.36TGSDD469 pKa = 3.91LIVTTTTYY477 pKa = 11.47GEE479 pKa = 4.37VPATYY484 pKa = 10.71NPVTDD489 pKa = 4.79TIRR492 pKa = 11.84RR493 pKa = 11.84DD494 pKa = 3.2PDD496 pKa = 3.13QPGFYY501 pKa = 9.9IMEE504 pKa = 4.64RR505 pKa = 11.84PLPSAQKK512 pKa = 10.47GLIYY516 pKa = 10.45FMNTTTFAHH525 pKa = 6.18IHH527 pKa = 5.34TVLKK531 pKa = 10.78AKK533 pKa = 10.4GVFQYY538 pKa = 11.41YY539 pKa = 10.36LYY541 pKa = 10.82ASSMDD546 pKa = 3.36QNVPSTSLKK555 pKa = 8.56YY556 pKa = 10.93SKK558 pKa = 10.17IIKK561 pKa = 8.53TNRR564 pKa = 11.84QRR566 pKa = 11.84RR567 pKa = 11.84AVNPSTVFYY576 pKa = 10.33NYY578 pKa = 7.21PTSAVNVPTNCPNIVLWAEE597 pKa = 4.01GLVATTSIYY606 pKa = 10.31QGCQRR611 pKa = 11.84EE612 pKa = 4.16NAASSLCWYY621 pKa = 8.77RR622 pKa = 11.84QSSFLCVPSTEE633 pKa = 4.05VTLITKK639 pKa = 6.91TTKK642 pKa = 9.57SCKK645 pKa = 10.26GINTNTLLPTSSASSVMCLDD665 pKa = 3.43LTLYY669 pKa = 6.7TTEE672 pKa = 4.53VSLCGNINAPTPTTKK687 pKa = 10.33VQLSLPSPDD696 pKa = 3.35SLTWGYY702 pKa = 11.34ASTPIYY708 pKa = 10.31SGYY711 pKa = 8.52LTHH714 pKa = 7.03TEE716 pKa = 4.49VISKK720 pKa = 10.21PPFCTSFTSLPSCAEE735 pKa = 4.54LICGSNSLCYY745 pKa = 9.77TKK747 pKa = 10.1LTEE750 pKa = 4.24FGKK753 pKa = 11.12LNMCTALGDD762 pKa = 4.46AISVYY767 pKa = 10.9NSVITDD773 pKa = 3.4IVEE776 pKa = 4.22SQATLDD782 pKa = 3.6ALYY785 pKa = 10.57TEE787 pKa = 4.64MLDD790 pKa = 3.64VAMTKK795 pKa = 10.54FGTVAQTRR803 pKa = 11.84KK804 pKa = 9.78KK805 pKa = 10.57RR806 pKa = 11.84FVEE809 pKa = 6.08FIALGVASVALVEE822 pKa = 4.14ATVAIGLAVANANDD836 pKa = 3.24IRR838 pKa = 11.84EE839 pKa = 4.09LRR841 pKa = 11.84GDD843 pKa = 3.73VKK845 pKa = 10.94NLRR848 pKa = 11.84DD849 pKa = 3.32EE850 pKa = 4.19SAANFKK856 pKa = 10.81AIEE859 pKa = 4.22EE860 pKa = 4.13KK861 pKa = 10.88HH862 pKa = 5.48NAFVKK867 pKa = 8.04TTADD871 pKa = 3.08GFAAQNEE878 pKa = 4.55VNKK881 pKa = 10.44GFNSNIDD888 pKa = 3.56TLQNSLKK895 pKa = 10.96DD896 pKa = 3.54MAIVTSQKK904 pKa = 10.33FKK906 pKa = 11.04EE907 pKa = 4.1LDD909 pKa = 3.21QAIQSLVSSINSLQLKK925 pKa = 10.22LDD927 pKa = 3.67TEE929 pKa = 4.58IQLSANSDD937 pKa = 3.51SALARR942 pKa = 11.84VLTALSLGHH951 pKa = 5.73RR952 pKa = 11.84TNRR955 pKa = 11.84NLDD958 pKa = 3.31RR959 pKa = 11.84HH960 pKa = 5.76ISQVRR965 pKa = 11.84TCLASIQQNSLAGCVNQGSNSIPLGIRR992 pKa = 11.84VFNEE996 pKa = 3.64TGDD999 pKa = 3.72LKK1001 pKa = 10.85IVFFYY1006 pKa = 9.87ATRR1009 pKa = 11.84QRR1011 pKa = 11.84LGYY1014 pKa = 9.6LQFKK1018 pKa = 8.64ATSAFCTNGTLVTAAPGCIFTANGTQIEE1046 pKa = 4.24HH1047 pKa = 6.35SKK1049 pKa = 10.34IGPLTPCGSSFVSLPIATCPTGSVDD1074 pKa = 3.55IEE1076 pKa = 4.4GVKK1079 pKa = 10.5PSLTPNFEE1087 pKa = 4.15VPQLEE1092 pKa = 4.46LKK1094 pKa = 10.19PPKK1097 pKa = 10.61LEE1099 pKa = 4.19VNLTNVNLSDD1109 pKa = 4.5YY1110 pKa = 10.03IRR1112 pKa = 11.84PITPVDD1118 pKa = 3.6LSKK1121 pKa = 10.85IEE1123 pKa = 4.31DD1124 pKa = 3.42LQARR1128 pKa = 11.84FDD1130 pKa = 4.06KK1131 pKa = 11.34LNLTLTEE1138 pKa = 4.43LSHH1141 pKa = 6.91ISSSGSGLPTWAIIFIVCVAVVIVLVFIVTIYY1173 pKa = 10.47MNTRR1177 pKa = 11.84QSTRR1181 pKa = 11.84LDD1183 pKa = 3.38SFLSKK1188 pKa = 10.31IRR1190 pKa = 3.6
MM1 pKa = 7.31MLLILPLILLCSSTHH16 pKa = 6.47AYY18 pKa = 9.16RR19 pKa = 11.84VQFVAPVPLNWINCYY34 pKa = 7.76TTDD37 pKa = 3.64CVSWQACPTTVAALVCVRR55 pKa = 11.84FTAVAPSFYY64 pKa = 10.8LSANNTLIGYY74 pKa = 8.4NADD77 pKa = 3.46PNTFYY82 pKa = 11.51DD83 pKa = 5.49LITMVQQSTYY93 pKa = 10.88SFSQAQTDD101 pKa = 3.92TPPTKK106 pKa = 10.18QCNGPSTILSGQCLTPADD124 pKa = 3.95SCLDD128 pKa = 3.86PLCPTTSFFEE138 pKa = 4.33TGSLDD143 pKa = 4.07SLTLDD148 pKa = 3.84TQCVASYY155 pKa = 10.32KK156 pKa = 10.64GYY158 pKa = 9.81GSVYY162 pKa = 9.03GTPCEE167 pKa = 4.02PFAPQLSPYY176 pKa = 7.91FTKK179 pKa = 10.68NVPNTPGFTDD189 pKa = 3.36YY190 pKa = 11.21GVNRR194 pKa = 11.84QSPKK198 pKa = 9.9LGCYY202 pKa = 7.25YY203 pKa = 10.21EE204 pKa = 4.71KK205 pKa = 9.75WAKK208 pKa = 9.84YY209 pKa = 9.42RR210 pKa = 11.84PKK212 pKa = 10.34PYY214 pKa = 9.56TATIDD219 pKa = 3.68SLAGCDD225 pKa = 2.8IVFFFVNTLSDD236 pKa = 3.11RR237 pKa = 11.84SGNMGVFEE245 pKa = 4.99RR246 pKa = 11.84EE247 pKa = 3.64EE248 pKa = 4.63GDD250 pKa = 3.32WNTIAAIKK258 pKa = 9.64TKK260 pKa = 10.47FPTTQVFLTFGGWTSDD276 pKa = 3.44NKK278 pKa = 10.52IMSAAMSNSTILNDD292 pKa = 3.06INTLSSRR299 pKa = 11.84LGVSVDD305 pKa = 4.06FDD307 pKa = 3.78IEE309 pKa = 4.51FPGSSNGNAAVFPYY323 pKa = 10.67DD324 pKa = 3.42EE325 pKa = 4.64ALITSFMTDD334 pKa = 1.69ICNFQHH340 pKa = 6.82SAGRR344 pKa = 11.84QCAWGGLSVGSHH356 pKa = 5.3WHH358 pKa = 5.49PTLINKK364 pKa = 9.75LSTIAGVDD372 pKa = 3.23YY373 pKa = 10.58FPIFGYY379 pKa = 10.34DD380 pKa = 3.3LHH382 pKa = 7.79GSWAPIPRR390 pKa = 11.84QQSALVNYY398 pKa = 10.27SPDD401 pKa = 3.45PTVSGGGPLNTYY413 pKa = 9.96SLITSVDD420 pKa = 3.2NFLTIVPASKK430 pKa = 10.71LILGLPMYY438 pKa = 10.83ARR440 pKa = 11.84GYY442 pKa = 10.78LVDD445 pKa = 4.01SSNSVLGSFPLTGPMLGPGEE465 pKa = 4.36TGSDD469 pKa = 3.91LIVTTTTYY477 pKa = 11.47GEE479 pKa = 4.37VPATYY484 pKa = 10.71NPVTDD489 pKa = 4.79TIRR492 pKa = 11.84RR493 pKa = 11.84DD494 pKa = 3.2PDD496 pKa = 3.13QPGFYY501 pKa = 9.9IMEE504 pKa = 4.64RR505 pKa = 11.84PLPSAQKK512 pKa = 10.47GLIYY516 pKa = 10.45FMNTTTFAHH525 pKa = 6.18IHH527 pKa = 5.34TVLKK531 pKa = 10.78AKK533 pKa = 10.4GVFQYY538 pKa = 11.41YY539 pKa = 10.36LYY541 pKa = 10.82ASSMDD546 pKa = 3.36QNVPSTSLKK555 pKa = 8.56YY556 pKa = 10.93SKK558 pKa = 10.17IIKK561 pKa = 8.53TNRR564 pKa = 11.84QRR566 pKa = 11.84RR567 pKa = 11.84AVNPSTVFYY576 pKa = 10.33NYY578 pKa = 7.21PTSAVNVPTNCPNIVLWAEE597 pKa = 4.01GLVATTSIYY606 pKa = 10.31QGCQRR611 pKa = 11.84EE612 pKa = 4.16NAASSLCWYY621 pKa = 8.77RR622 pKa = 11.84QSSFLCVPSTEE633 pKa = 4.05VTLITKK639 pKa = 6.91TTKK642 pKa = 9.57SCKK645 pKa = 10.26GINTNTLLPTSSASSVMCLDD665 pKa = 3.43LTLYY669 pKa = 6.7TTEE672 pKa = 4.53VSLCGNINAPTPTTKK687 pKa = 10.33VQLSLPSPDD696 pKa = 3.35SLTWGYY702 pKa = 11.34ASTPIYY708 pKa = 10.31SGYY711 pKa = 8.52LTHH714 pKa = 7.03TEE716 pKa = 4.49VISKK720 pKa = 10.21PPFCTSFTSLPSCAEE735 pKa = 4.54LICGSNSLCYY745 pKa = 9.77TKK747 pKa = 10.1LTEE750 pKa = 4.24FGKK753 pKa = 11.12LNMCTALGDD762 pKa = 4.46AISVYY767 pKa = 10.9NSVITDD773 pKa = 3.4IVEE776 pKa = 4.22SQATLDD782 pKa = 3.6ALYY785 pKa = 10.57TEE787 pKa = 4.64MLDD790 pKa = 3.64VAMTKK795 pKa = 10.54FGTVAQTRR803 pKa = 11.84KK804 pKa = 9.78KK805 pKa = 10.57RR806 pKa = 11.84FVEE809 pKa = 6.08FIALGVASVALVEE822 pKa = 4.14ATVAIGLAVANANDD836 pKa = 3.24IRR838 pKa = 11.84EE839 pKa = 4.09LRR841 pKa = 11.84GDD843 pKa = 3.73VKK845 pKa = 10.94NLRR848 pKa = 11.84DD849 pKa = 3.32EE850 pKa = 4.19SAANFKK856 pKa = 10.81AIEE859 pKa = 4.22EE860 pKa = 4.13KK861 pKa = 10.88HH862 pKa = 5.48NAFVKK867 pKa = 8.04TTADD871 pKa = 3.08GFAAQNEE878 pKa = 4.55VNKK881 pKa = 10.44GFNSNIDD888 pKa = 3.56TLQNSLKK895 pKa = 10.96DD896 pKa = 3.54MAIVTSQKK904 pKa = 10.33FKK906 pKa = 11.04EE907 pKa = 4.1LDD909 pKa = 3.21QAIQSLVSSINSLQLKK925 pKa = 10.22LDD927 pKa = 3.67TEE929 pKa = 4.58IQLSANSDD937 pKa = 3.51SALARR942 pKa = 11.84VLTALSLGHH951 pKa = 5.73RR952 pKa = 11.84TNRR955 pKa = 11.84NLDD958 pKa = 3.31RR959 pKa = 11.84HH960 pKa = 5.76ISQVRR965 pKa = 11.84TCLASIQQNSLAGCVNQGSNSIPLGIRR992 pKa = 11.84VFNEE996 pKa = 3.64TGDD999 pKa = 3.72LKK1001 pKa = 10.85IVFFYY1006 pKa = 9.87ATRR1009 pKa = 11.84QRR1011 pKa = 11.84LGYY1014 pKa = 9.6LQFKK1018 pKa = 8.64ATSAFCTNGTLVTAAPGCIFTANGTQIEE1046 pKa = 4.24HH1047 pKa = 6.35SKK1049 pKa = 10.34IGPLTPCGSSFVSLPIATCPTGSVDD1074 pKa = 3.55IEE1076 pKa = 4.4GVKK1079 pKa = 10.5PSLTPNFEE1087 pKa = 4.15VPQLEE1092 pKa = 4.46LKK1094 pKa = 10.19PPKK1097 pKa = 10.61LEE1099 pKa = 4.19VNLTNVNLSDD1109 pKa = 4.5YY1110 pKa = 10.03IRR1112 pKa = 11.84PITPVDD1118 pKa = 3.6LSKK1121 pKa = 10.85IEE1123 pKa = 4.31DD1124 pKa = 3.42LQARR1128 pKa = 11.84FDD1130 pKa = 4.06KK1131 pKa = 11.34LNLTLTEE1138 pKa = 4.43LSHH1141 pKa = 6.91ISSSGSGLPTWAIIFIVCVAVVIVLVFIVTIYY1173 pKa = 10.47MNTRR1177 pKa = 11.84QSTRR1181 pKa = 11.84LDD1183 pKa = 3.38SFLSKK1188 pKa = 10.31IRR1190 pKa = 3.6
Molecular weight: 129.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I6LMN3|I6LMN3_9NIDO Nucleocapsid protein OS=Fathead minnow nidovirus OX=889873 GN=N PE=4 SV=1
MM1 pKa = 7.56SFPMQPAYY9 pKa = 10.47FPLQQRR15 pKa = 11.84QRR17 pKa = 11.84RR18 pKa = 11.84PRR20 pKa = 11.84AVAGAKK26 pKa = 9.84PKK28 pKa = 10.42FKK30 pKa = 10.52SAPPSPAVINAAVKK44 pKa = 10.64KK45 pKa = 9.73EE46 pKa = 4.06VSTHH50 pKa = 5.57IPMPPISMQEE60 pKa = 3.87KK61 pKa = 9.61SGLRR65 pKa = 11.84ALDD68 pKa = 3.19VRR70 pKa = 11.84NNFSTAEE77 pKa = 3.98LAGLYY82 pKa = 10.01AQLIQHH88 pKa = 7.18LNHH91 pKa = 6.08CHH93 pKa = 4.82GRR95 pKa = 11.84MVFMPGDD102 pKa = 4.08TISQGFVQVQLKK114 pKa = 10.33LRR116 pKa = 11.84LPSDD120 pKa = 3.29QCLKK124 pKa = 10.04MVSAHH129 pKa = 6.17SFAEE133 pKa = 3.87QRR135 pKa = 11.84LLASEE140 pKa = 5.04GEE142 pKa = 4.22ASVTTTLLSSLDD154 pKa = 3.66KK155 pKa = 11.05QAVNSLNAKK164 pKa = 9.89DD165 pKa = 3.42
MM1 pKa = 7.56SFPMQPAYY9 pKa = 10.47FPLQQRR15 pKa = 11.84QRR17 pKa = 11.84RR18 pKa = 11.84PRR20 pKa = 11.84AVAGAKK26 pKa = 9.84PKK28 pKa = 10.42FKK30 pKa = 10.52SAPPSPAVINAAVKK44 pKa = 10.64KK45 pKa = 9.73EE46 pKa = 4.06VSTHH50 pKa = 5.57IPMPPISMQEE60 pKa = 3.87KK61 pKa = 9.61SGLRR65 pKa = 11.84ALDD68 pKa = 3.19VRR70 pKa = 11.84NNFSTAEE77 pKa = 3.98LAGLYY82 pKa = 10.01AQLIQHH88 pKa = 7.18LNHH91 pKa = 6.08CHH93 pKa = 4.82GRR95 pKa = 11.84MVFMPGDD102 pKa = 4.08TISQGFVQVQLKK114 pKa = 10.33LRR116 pKa = 11.84LPSDD120 pKa = 3.29QCLKK124 pKa = 10.04MVSAHH129 pKa = 6.17SFAEE133 pKa = 3.87QRR135 pKa = 11.84LLASEE140 pKa = 5.04GEE142 pKa = 4.22ASVTTTLLSSLDD154 pKa = 3.66KK155 pKa = 11.05QAVNSLNAKK164 pKa = 9.89DD165 pKa = 3.42
Molecular weight: 18.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13614 |
165 |
7174 |
2722.8 |
304.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.847 ± 0.515 | 1.814 ± 0.192 |
4.348 ± 0.19 | 4.415 ± 0.41 |
4.606 ± 0.102 | 4.26 ± 0.333 |
3.012 ± 0.371 | 6.339 ± 0.22 |
5.406 ± 0.361 | 9.079 ± 0.363 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.035 ± 0.085 | 5.362 ± 0.124 |
5.428 ± 0.191 | 4.356 ± 0.169 |
2.637 ± 0.187 | 8.219 ± 0.336 |
10.842 ± 0.594 | 6.846 ± 0.172 |
0.654 ± 0.062 | 4.495 ± 0.281 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
