
Wallal virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|U3RCS2|U3RCS2_9REOV Outer capsid protein VP2 OS=Wallal virus OX=40061 PE=3 SV=1
MM1 pKa = 7.17EE2 pKa = 4.63QRR4 pKa = 11.84TQLRR8 pKa = 11.84RR9 pKa = 11.84VFPKK13 pKa = 9.44TVCVLDD19 pKa = 3.98PKK21 pKa = 11.37GEE23 pKa = 4.49TICGMIAKK31 pKa = 9.04SQHH34 pKa = 5.8KK35 pKa = 9.11PYY37 pKa = 10.63CLVRR41 pKa = 11.84TGRR44 pKa = 11.84VICLKK49 pKa = 10.45AISQPPPKK57 pKa = 10.25AYY59 pKa = 10.07VLEE62 pKa = 4.32VVKK65 pKa = 10.86CGAYY69 pKa = 10.14RR70 pKa = 11.84LIDD73 pKa = 4.01GQDD76 pKa = 3.32QISIMLNKK84 pKa = 10.22DD85 pKa = 3.53CVEE88 pKa = 4.14MTSEE92 pKa = 4.78RR93 pKa = 11.84WEE95 pKa = 3.78EE96 pKa = 3.64WQYY99 pKa = 11.89EE100 pKa = 4.32MVSIKK105 pKa = 10.64PMVVSLQLEE114 pKa = 4.47YY115 pKa = 11.16GVVDD119 pKa = 4.52AEE121 pKa = 4.24LKK123 pKa = 9.0YY124 pKa = 10.91GKK126 pKa = 10.43GFGSVEE132 pKa = 3.96PYY134 pKa = 10.32KK135 pKa = 10.94KK136 pKa = 10.35NEE138 pKa = 4.01LEE140 pKa = 4.12RR141 pKa = 11.84GEE143 pKa = 4.86VPTLPGVQQSAVGLRR158 pKa = 11.84EE159 pKa = 4.19LRR161 pKa = 11.84TKK163 pKa = 10.43LKK165 pKa = 10.5KK166 pKa = 10.0EE167 pKa = 3.96RR168 pKa = 11.84EE169 pKa = 4.12EE170 pKa = 4.09KK171 pKa = 9.34VQGRR175 pKa = 11.84SVEE178 pKa = 4.12LPTMKK183 pKa = 10.22KK184 pKa = 10.56VNVQAEE190 pKa = 4.21QGFEE194 pKa = 3.52WSAVRR199 pKa = 11.84EE200 pKa = 4.23KK201 pKa = 10.62VVEE204 pKa = 4.03MEE206 pKa = 4.74EE207 pKa = 4.49EE208 pKa = 3.88IKK210 pKa = 10.76DD211 pKa = 3.65WAEE214 pKa = 4.05HH215 pKa = 5.94GSSEE219 pKa = 5.14EE220 pKa = 4.17EE221 pKa = 4.27INDD224 pKa = 3.58KK225 pKa = 10.62QQTDD229 pKa = 3.63DD230 pKa = 4.01EE231 pKa = 4.83EE232 pKa = 4.64VVKK235 pKa = 11.03DD236 pKa = 4.34DD237 pKa = 5.03DD238 pKa = 5.61DD239 pKa = 3.96EE240 pKa = 4.42ATEE243 pKa = 4.11EE244 pKa = 4.36GLVKK248 pKa = 10.26VDD250 pKa = 3.92SFITGDD256 pKa = 3.51YY257 pKa = 9.61VNYY260 pKa = 9.92VSGMSKK266 pKa = 10.48KK267 pKa = 9.99KK268 pKa = 10.28DD269 pKa = 3.41EE270 pKa = 4.42RR271 pKa = 11.84LSGLSLSFPKK281 pKa = 10.17IEE283 pKa = 4.27GEE285 pKa = 4.13FEE287 pKa = 3.82KK288 pKa = 11.0VLCIKK293 pKa = 9.73KK294 pKa = 8.42TEE296 pKa = 4.06QTGFPVFEE304 pKa = 4.19VDD306 pKa = 3.67QATKK310 pKa = 9.09TFRR313 pKa = 11.84FRR315 pKa = 11.84MIGKK319 pKa = 8.62CDD321 pKa = 2.96RR322 pKa = 11.84VFVVKK327 pKa = 10.33RR328 pKa = 11.84SMSMVYY334 pKa = 10.21LPAGGPLL341 pKa = 3.3
MM1 pKa = 7.17EE2 pKa = 4.63QRR4 pKa = 11.84TQLRR8 pKa = 11.84RR9 pKa = 11.84VFPKK13 pKa = 9.44TVCVLDD19 pKa = 3.98PKK21 pKa = 11.37GEE23 pKa = 4.49TICGMIAKK31 pKa = 9.04SQHH34 pKa = 5.8KK35 pKa = 9.11PYY37 pKa = 10.63CLVRR41 pKa = 11.84TGRR44 pKa = 11.84VICLKK49 pKa = 10.45AISQPPPKK57 pKa = 10.25AYY59 pKa = 10.07VLEE62 pKa = 4.32VVKK65 pKa = 10.86CGAYY69 pKa = 10.14RR70 pKa = 11.84LIDD73 pKa = 4.01GQDD76 pKa = 3.32QISIMLNKK84 pKa = 10.22DD85 pKa = 3.53CVEE88 pKa = 4.14MTSEE92 pKa = 4.78RR93 pKa = 11.84WEE95 pKa = 3.78EE96 pKa = 3.64WQYY99 pKa = 11.89EE100 pKa = 4.32MVSIKK105 pKa = 10.64PMVVSLQLEE114 pKa = 4.47YY115 pKa = 11.16GVVDD119 pKa = 4.52AEE121 pKa = 4.24LKK123 pKa = 9.0YY124 pKa = 10.91GKK126 pKa = 10.43GFGSVEE132 pKa = 3.96PYY134 pKa = 10.32KK135 pKa = 10.94KK136 pKa = 10.35NEE138 pKa = 4.01LEE140 pKa = 4.12RR141 pKa = 11.84GEE143 pKa = 4.86VPTLPGVQQSAVGLRR158 pKa = 11.84EE159 pKa = 4.19LRR161 pKa = 11.84TKK163 pKa = 10.43LKK165 pKa = 10.5KK166 pKa = 10.0EE167 pKa = 3.96RR168 pKa = 11.84EE169 pKa = 4.12EE170 pKa = 4.09KK171 pKa = 9.34VQGRR175 pKa = 11.84SVEE178 pKa = 4.12LPTMKK183 pKa = 10.22KK184 pKa = 10.56VNVQAEE190 pKa = 4.21QGFEE194 pKa = 3.52WSAVRR199 pKa = 11.84EE200 pKa = 4.23KK201 pKa = 10.62VVEE204 pKa = 4.03MEE206 pKa = 4.74EE207 pKa = 4.49EE208 pKa = 3.88IKK210 pKa = 10.76DD211 pKa = 3.65WAEE214 pKa = 4.05HH215 pKa = 5.94GSSEE219 pKa = 5.14EE220 pKa = 4.17EE221 pKa = 4.27INDD224 pKa = 3.58KK225 pKa = 10.62QQTDD229 pKa = 3.63DD230 pKa = 4.01EE231 pKa = 4.83EE232 pKa = 4.64VVKK235 pKa = 11.03DD236 pKa = 4.34DD237 pKa = 5.03DD238 pKa = 5.61DD239 pKa = 3.96EE240 pKa = 4.42ATEE243 pKa = 4.11EE244 pKa = 4.36GLVKK248 pKa = 10.26VDD250 pKa = 3.92SFITGDD256 pKa = 3.51YY257 pKa = 9.61VNYY260 pKa = 9.92VSGMSKK266 pKa = 10.48KK267 pKa = 9.99KK268 pKa = 10.28DD269 pKa = 3.41EE270 pKa = 4.42RR271 pKa = 11.84LSGLSLSFPKK281 pKa = 10.17IEE283 pKa = 4.27GEE285 pKa = 4.13FEE287 pKa = 3.82KK288 pKa = 11.0VLCIKK293 pKa = 9.73KK294 pKa = 8.42TEE296 pKa = 4.06QTGFPVFEE304 pKa = 4.19VDD306 pKa = 3.67QATKK310 pKa = 9.09TFRR313 pKa = 11.84FRR315 pKa = 11.84MIGKK319 pKa = 8.62CDD321 pKa = 2.96RR322 pKa = 11.84VFVVKK327 pKa = 10.33RR328 pKa = 11.84SMSMVYY334 pKa = 10.21LPAGGPLL341 pKa = 3.3
Molecular weight: 38.86 kDa
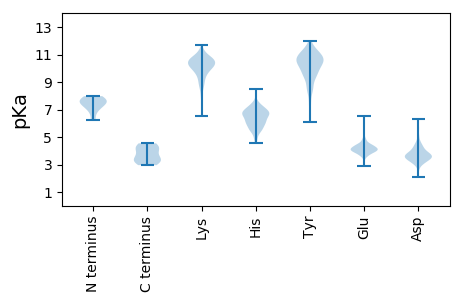
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U3RHX5|U3RHX5_9REOV RNA-directed RNA polymerase OS=Wallal virus OX=40061 PE=3 SV=1
MM1 pKa = 7.56LSEE4 pKa = 3.89LAARR8 pKa = 11.84FEE10 pKa = 4.53SEE12 pKa = 3.97KK13 pKa = 10.4VRR15 pKa = 11.84YY16 pKa = 9.15SATEE20 pKa = 3.44QQLITKK26 pKa = 9.37EE27 pKa = 4.09EE28 pKa = 4.03EE29 pKa = 3.71EE30 pKa = 4.66PIYY33 pKa = 11.15EE34 pKa = 4.26RR35 pKa = 11.84MDD37 pKa = 3.63KK38 pKa = 11.05AVVPYY43 pKa = 10.1SRR45 pKa = 11.84PPSYY49 pKa = 11.25VPTAPSGVSGKK60 pKa = 10.15DD61 pKa = 3.18VSLDD65 pKa = 3.23ILNNAMSNTTGATNAMKK82 pKa = 9.8MEE84 pKa = 3.92KK85 pKa = 8.9TAYY88 pKa = 9.82SSYY91 pKa = 11.01AEE93 pKa = 4.15AMRR96 pKa = 11.84DD97 pKa = 3.57SVPVQNVKK105 pKa = 9.99TSVNALIIPRR115 pKa = 11.84LRR117 pKa = 11.84NEE119 pKa = 3.7LSGLKK124 pKa = 9.8RR125 pKa = 11.84KK126 pKa = 8.23RR127 pKa = 11.84TLVHH131 pKa = 6.06IALIVTAAVTMITSFSSLVKK151 pKa = 10.27DD152 pKa = 3.99FQVSIPSGTEE162 pKa = 3.62SGNSTVTVQVPKK174 pKa = 10.13WFASFSAVFNTVNLVATGLMISFARR199 pKa = 11.84MEE201 pKa = 4.3KK202 pKa = 9.87MLDD205 pKa = 3.48GQIGMIKK212 pKa = 9.98KK213 pKa = 10.15EE214 pKa = 3.9IMKK217 pKa = 10.35KK218 pKa = 9.4EE219 pKa = 4.22SYY221 pKa = 10.73NEE223 pKa = 3.85AVRR226 pKa = 11.84KK227 pKa = 9.57SVTAIDD233 pKa = 3.75MTSLFQEE240 pKa = 4.32AEE242 pKa = 4.21GANGG246 pKa = 3.23
MM1 pKa = 7.56LSEE4 pKa = 3.89LAARR8 pKa = 11.84FEE10 pKa = 4.53SEE12 pKa = 3.97KK13 pKa = 10.4VRR15 pKa = 11.84YY16 pKa = 9.15SATEE20 pKa = 3.44QQLITKK26 pKa = 9.37EE27 pKa = 4.09EE28 pKa = 4.03EE29 pKa = 3.71EE30 pKa = 4.66PIYY33 pKa = 11.15EE34 pKa = 4.26RR35 pKa = 11.84MDD37 pKa = 3.63KK38 pKa = 11.05AVVPYY43 pKa = 10.1SRR45 pKa = 11.84PPSYY49 pKa = 11.25VPTAPSGVSGKK60 pKa = 10.15DD61 pKa = 3.18VSLDD65 pKa = 3.23ILNNAMSNTTGATNAMKK82 pKa = 9.8MEE84 pKa = 3.92KK85 pKa = 8.9TAYY88 pKa = 9.82SSYY91 pKa = 11.01AEE93 pKa = 4.15AMRR96 pKa = 11.84DD97 pKa = 3.57SVPVQNVKK105 pKa = 9.99TSVNALIIPRR115 pKa = 11.84LRR117 pKa = 11.84NEE119 pKa = 3.7LSGLKK124 pKa = 9.8RR125 pKa = 11.84KK126 pKa = 8.23RR127 pKa = 11.84TLVHH131 pKa = 6.06IALIVTAAVTMITSFSSLVKK151 pKa = 10.27DD152 pKa = 3.99FQVSIPSGTEE162 pKa = 3.62SGNSTVTVQVPKK174 pKa = 10.13WFASFSAVFNTVNLVATGLMISFARR199 pKa = 11.84MEE201 pKa = 4.3KK202 pKa = 9.87MLDD205 pKa = 3.48GQIGMIKK212 pKa = 9.98KK213 pKa = 10.15EE214 pKa = 3.9IMKK217 pKa = 10.35KK218 pKa = 9.4EE219 pKa = 4.22SYY221 pKa = 10.73NEE223 pKa = 3.85AVRR226 pKa = 11.84KK227 pKa = 9.57SVTAIDD233 pKa = 3.75MTSLFQEE240 pKa = 4.32AEE242 pKa = 4.21GANGG246 pKa = 3.23
Molecular weight: 26.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6186 |
246 |
1306 |
618.6 |
71.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.24 ± 0.527 | 1.099 ± 0.285 |
5.706 ± 0.285 | 7.646 ± 0.528 |
3.831 ± 0.235 | 5.48 ± 0.381 |
2.069 ± 0.227 | 6.579 ± 0.435 |
6.531 ± 0.787 | 8.697 ± 0.277 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.767 ± 0.309 | 4.64 ± 0.255 |
3.831 ± 0.332 | 3.686 ± 0.31 |
6.466 ± 0.336 | 5.674 ± 0.369 |
5.318 ± 0.373 | 7.355 ± 0.363 |
1.406 ± 0.265 | 3.977 ± 0.207 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
