
Sellimonas intestinalis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Sellimonas
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
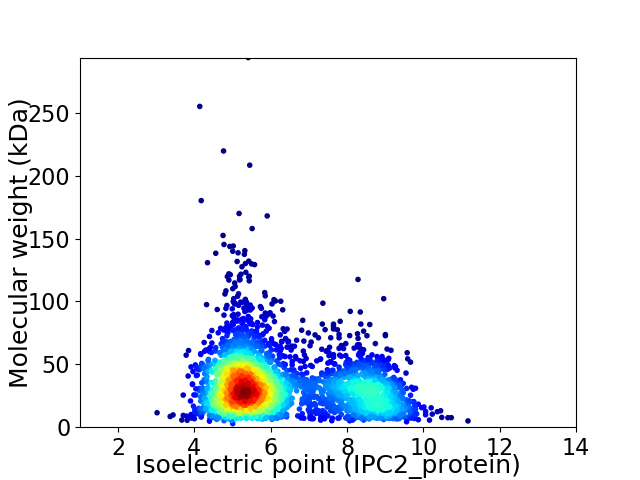
Virtual 2D-PAGE plot for 3054 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3E3JZG1|A0A3E3JZG1_9FIRM Peptidase M23 OS=Sellimonas intestinalis OX=1653434 GN=DW016_14435 PE=4 SV=1
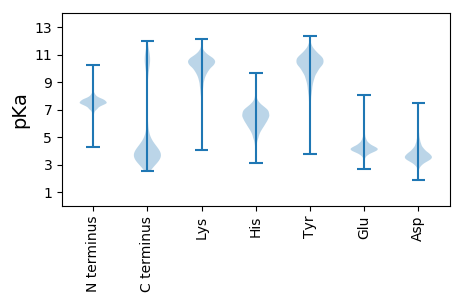
MM1 pKa = 7.88RR2 pKa = 11.84KK3 pKa = 8.84GKK5 pKa = 10.26KK6 pKa = 10.08VIGAALAGCMTASMFLTACGDD27 pKa = 3.82GNEE30 pKa = 4.48KK31 pKa = 10.43SGKK34 pKa = 7.58TEE36 pKa = 4.39IEE38 pKa = 3.59ILQYY42 pKa = 10.67KK43 pKa = 9.43PEE45 pKa = 4.15AATYY49 pKa = 9.68FDD51 pKa = 3.65QVEE54 pKa = 4.29EE55 pKa = 4.52KK56 pKa = 10.94FNATHH61 pKa = 7.52DD62 pKa = 5.1DD63 pKa = 2.92IHH65 pKa = 6.34LTIDD69 pKa = 3.73SPNDD73 pKa = 2.87ASTIMRR79 pKa = 11.84TRR81 pKa = 11.84FIRR84 pKa = 11.84EE85 pKa = 4.35DD86 pKa = 3.57YY87 pKa = 10.57PDD89 pKa = 4.29IIGIGGDD96 pKa = 3.25INYY99 pKa = 10.09SYY101 pKa = 11.41YY102 pKa = 10.86VDD104 pKa = 4.11AGILADD110 pKa = 3.77VSDD113 pKa = 4.5SEE115 pKa = 4.83VMGEE119 pKa = 4.33IKK121 pKa = 10.5DD122 pKa = 3.77SYY124 pKa = 11.35KK125 pKa = 10.96DD126 pKa = 3.13ILEE129 pKa = 4.25ALEE132 pKa = 3.97IVPTEE137 pKa = 4.01GTFGIPYY144 pKa = 8.54VANAAGILYY153 pKa = 9.69NRR155 pKa = 11.84DD156 pKa = 3.01MFEE159 pKa = 3.74EE160 pKa = 4.54HH161 pKa = 6.39GWEE164 pKa = 5.12IPNTWDD170 pKa = 4.82EE171 pKa = 5.93LMTLCNDD178 pKa = 3.25IQSEE182 pKa = 4.85GILPFYY188 pKa = 10.77FGFKK192 pKa = 9.56DD193 pKa = 2.9TWTCLAPWNSMAVALAPPDD212 pKa = 3.34LCKK215 pKa = 10.23QVNKK219 pKa = 10.47GDD221 pKa = 4.05ANFTDD226 pKa = 4.37SYY228 pKa = 11.39RR229 pKa = 11.84EE230 pKa = 3.79VAEE233 pKa = 5.12KK234 pKa = 10.81YY235 pKa = 10.73LEE237 pKa = 4.06LLNYY241 pKa = 10.11GPEE244 pKa = 4.05DD245 pKa = 3.66PFAYY249 pKa = 9.9GYY251 pKa = 11.37NDD253 pKa = 3.42ACTAFANGEE262 pKa = 4.09SAMYY266 pKa = 9.44PIGSYY271 pKa = 10.32AVPQILSVNPDD282 pKa = 3.23MNIDD286 pKa = 3.84SFVTPASDD294 pKa = 3.16TKK296 pKa = 10.97EE297 pKa = 4.07GNTLNSGVDD306 pKa = 3.29LMFAVTADD314 pKa = 4.06CEE316 pKa = 4.44NKK318 pKa = 8.86EE319 pKa = 3.75AAYY322 pKa = 9.88EE323 pKa = 4.0VLEE326 pKa = 4.27FLYY329 pKa = 10.67EE330 pKa = 4.13DD331 pKa = 4.49EE332 pKa = 5.58NIQAYY337 pKa = 9.68IDD339 pKa = 4.0DD340 pKa = 4.28QNAIPCKK347 pKa = 10.56EE348 pKa = 3.8GDD350 pKa = 3.74FDD352 pKa = 6.05LASQLDD358 pKa = 3.84GMTEE362 pKa = 4.03YY363 pKa = 10.49IEE365 pKa = 4.48AGNMTDD371 pKa = 3.89YY372 pKa = 10.93QDD374 pKa = 3.86HH375 pKa = 6.74YY376 pKa = 9.86YY377 pKa = 9.24PSEE380 pKa = 4.04MAVDD384 pKa = 3.86AQIQTFLINKK394 pKa = 9.55DD395 pKa = 2.41VDD397 pKa = 3.63AFLKK401 pKa = 10.85KK402 pKa = 10.15FDD404 pKa = 4.71SDD406 pKa = 3.08WTRR409 pKa = 11.84YY410 pKa = 9.06NRR412 pKa = 11.84DD413 pKa = 3.08IIRR416 pKa = 11.84EE417 pKa = 3.98VQEE420 pKa = 4.09YY421 pKa = 10.71EE422 pKa = 4.43EE423 pKa = 4.61EE424 pKa = 4.25NGDD427 pKa = 3.53AA428 pKa = 5.2
MM1 pKa = 7.88RR2 pKa = 11.84KK3 pKa = 8.84GKK5 pKa = 10.26KK6 pKa = 10.08VIGAALAGCMTASMFLTACGDD27 pKa = 3.82GNEE30 pKa = 4.48KK31 pKa = 10.43SGKK34 pKa = 7.58TEE36 pKa = 4.39IEE38 pKa = 3.59ILQYY42 pKa = 10.67KK43 pKa = 9.43PEE45 pKa = 4.15AATYY49 pKa = 9.68FDD51 pKa = 3.65QVEE54 pKa = 4.29EE55 pKa = 4.52KK56 pKa = 10.94FNATHH61 pKa = 7.52DD62 pKa = 5.1DD63 pKa = 2.92IHH65 pKa = 6.34LTIDD69 pKa = 3.73SPNDD73 pKa = 2.87ASTIMRR79 pKa = 11.84TRR81 pKa = 11.84FIRR84 pKa = 11.84EE85 pKa = 4.35DD86 pKa = 3.57YY87 pKa = 10.57PDD89 pKa = 4.29IIGIGGDD96 pKa = 3.25INYY99 pKa = 10.09SYY101 pKa = 11.41YY102 pKa = 10.86VDD104 pKa = 4.11AGILADD110 pKa = 3.77VSDD113 pKa = 4.5SEE115 pKa = 4.83VMGEE119 pKa = 4.33IKK121 pKa = 10.5DD122 pKa = 3.77SYY124 pKa = 11.35KK125 pKa = 10.96DD126 pKa = 3.13ILEE129 pKa = 4.25ALEE132 pKa = 3.97IVPTEE137 pKa = 4.01GTFGIPYY144 pKa = 8.54VANAAGILYY153 pKa = 9.69NRR155 pKa = 11.84DD156 pKa = 3.01MFEE159 pKa = 3.74EE160 pKa = 4.54HH161 pKa = 6.39GWEE164 pKa = 5.12IPNTWDD170 pKa = 4.82EE171 pKa = 5.93LMTLCNDD178 pKa = 3.25IQSEE182 pKa = 4.85GILPFYY188 pKa = 10.77FGFKK192 pKa = 9.56DD193 pKa = 2.9TWTCLAPWNSMAVALAPPDD212 pKa = 3.34LCKK215 pKa = 10.23QVNKK219 pKa = 10.47GDD221 pKa = 4.05ANFTDD226 pKa = 4.37SYY228 pKa = 11.39RR229 pKa = 11.84EE230 pKa = 3.79VAEE233 pKa = 5.12KK234 pKa = 10.81YY235 pKa = 10.73LEE237 pKa = 4.06LLNYY241 pKa = 10.11GPEE244 pKa = 4.05DD245 pKa = 3.66PFAYY249 pKa = 9.9GYY251 pKa = 11.37NDD253 pKa = 3.42ACTAFANGEE262 pKa = 4.09SAMYY266 pKa = 9.44PIGSYY271 pKa = 10.32AVPQILSVNPDD282 pKa = 3.23MNIDD286 pKa = 3.84SFVTPASDD294 pKa = 3.16TKK296 pKa = 10.97EE297 pKa = 4.07GNTLNSGVDD306 pKa = 3.29LMFAVTADD314 pKa = 4.06CEE316 pKa = 4.44NKK318 pKa = 8.86EE319 pKa = 3.75AAYY322 pKa = 9.88EE323 pKa = 4.0VLEE326 pKa = 4.27FLYY329 pKa = 10.67EE330 pKa = 4.13DD331 pKa = 4.49EE332 pKa = 5.58NIQAYY337 pKa = 9.68IDD339 pKa = 4.0DD340 pKa = 4.28QNAIPCKK347 pKa = 10.56EE348 pKa = 3.8GDD350 pKa = 3.74FDD352 pKa = 6.05LASQLDD358 pKa = 3.84GMTEE362 pKa = 4.03YY363 pKa = 10.49IEE365 pKa = 4.48AGNMTDD371 pKa = 3.89YY372 pKa = 10.93QDD374 pKa = 3.86HH375 pKa = 6.74YY376 pKa = 9.86YY377 pKa = 9.24PSEE380 pKa = 4.04MAVDD384 pKa = 3.86AQIQTFLINKK394 pKa = 9.55DD395 pKa = 2.41VDD397 pKa = 3.63AFLKK401 pKa = 10.85KK402 pKa = 10.15FDD404 pKa = 4.71SDD406 pKa = 3.08WTRR409 pKa = 11.84YY410 pKa = 9.06NRR412 pKa = 11.84DD413 pKa = 3.08IIRR416 pKa = 11.84EE417 pKa = 3.98VQEE420 pKa = 4.09YY421 pKa = 10.71EE422 pKa = 4.43EE423 pKa = 4.61EE424 pKa = 4.25NGDD427 pKa = 3.53AA428 pKa = 5.2
Molecular weight: 47.98 kDa
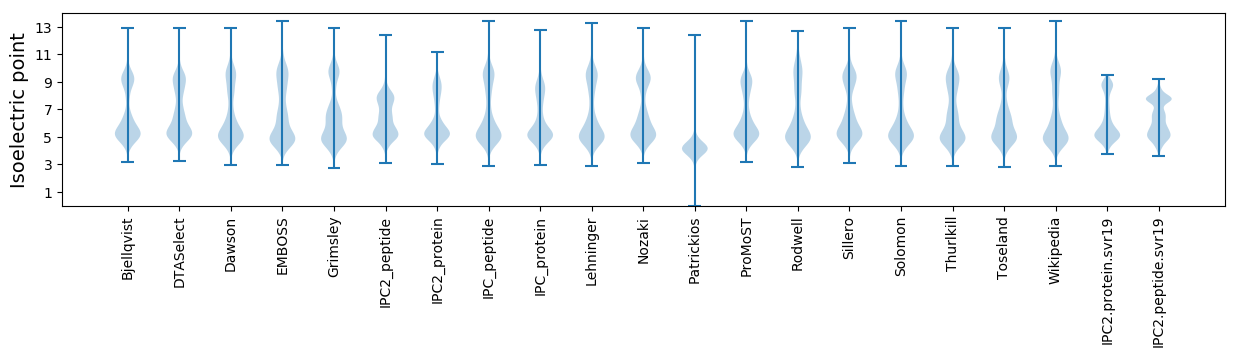
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3E3K0I5|A0A3E3K0I5_9FIRM tRNA uridine 5-carboxymethylaminomethyl modification enzyme MnmG OS=Sellimonas intestinalis OX=1653434 GN=mnmG PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 4.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
956250 |
25 |
2557 |
313.1 |
35.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.083 ± 0.046 | 1.496 ± 0.02 |
5.373 ± 0.034 | 8.039 ± 0.05 |
4.205 ± 0.032 | 7.243 ± 0.04 |
1.857 ± 0.019 | 7.379 ± 0.051 |
6.806 ± 0.035 | 9.074 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.175 ± 0.022 | 3.844 ± 0.031 |
3.399 ± 0.026 | 3.339 ± 0.027 |
4.955 ± 0.038 | 5.54 ± 0.037 |
5.301 ± 0.035 | 6.823 ± 0.037 |
0.98 ± 0.017 | 4.089 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
