
Turkey aviadenovirus 4
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Aviadenovirus; Turkey aviadenovirus C
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
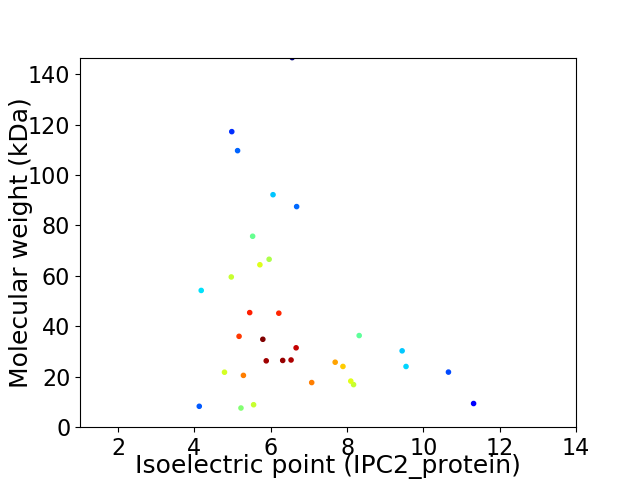
Virtual 2D-PAGE plot for 33 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U5NHL0|U5NHL0_9ADEN ORF8 OS=Turkey aviadenovirus 4 OX=1408257 PE=4 SV=1
MM1 pKa = 7.51SSAPFHH7 pKa = 6.34GMSISGRR14 pKa = 11.84KK15 pKa = 9.14RR16 pKa = 11.84SSNASNDD23 pKa = 2.91EE24 pKa = 4.15AEE26 pKa = 4.42PVKK29 pKa = 10.72APKK32 pKa = 9.81LQAGARR38 pKa = 11.84VTINAQNAEE47 pKa = 4.35LDD49 pKa = 3.61LVYY52 pKa = 10.23PFWYY56 pKa = 9.92EE57 pKa = 3.97STSTGGNPSVNPPLLDD73 pKa = 3.61PSGPLYY79 pKa = 10.46VQSNMLKK86 pKa = 9.83VRR88 pKa = 11.84TAAPVEE94 pKa = 4.2VANGSITLAYY104 pKa = 10.41DD105 pKa = 3.33DD106 pKa = 5.01TLTLDD111 pKa = 4.58DD112 pKa = 4.52QNKK115 pKa = 9.39LQINVEE121 pKa = 4.15PNGPLRR127 pKa = 11.84STPDD131 pKa = 3.54GLDD134 pKa = 3.33LAIDD138 pKa = 3.66STLEE142 pKa = 3.58VDD144 pKa = 3.05NWEE147 pKa = 4.85LGVKK151 pKa = 10.0LNPAGPIDD159 pKa = 4.01ASAAGLDD166 pKa = 3.6INVDD170 pKa = 3.42EE171 pKa = 4.61TLLVAEE177 pKa = 5.32DD178 pKa = 3.9STNQRR183 pKa = 11.84YY184 pKa = 8.55EE185 pKa = 4.22LGVHH189 pKa = 6.76LNPNGPITADD199 pKa = 3.23SDD201 pKa = 4.26GLDD204 pKa = 3.85LEE206 pKa = 5.04INPEE210 pKa = 4.17TLTVTNDD217 pKa = 3.11SNTGGVLSVVLKK229 pKa = 9.94PQGGLQSSLLGIGVAVDD246 pKa = 3.38RR247 pKa = 11.84TLTIDD252 pKa = 3.31QNTVEE257 pKa = 4.4VKK259 pKa = 10.08IDD261 pKa = 3.36PTGPLTSSDD270 pKa = 3.46NGLSLDD276 pKa = 3.71YY277 pKa = 9.8DD278 pKa = 3.8TTDD281 pKa = 3.83FSVQAGKK288 pKa = 10.8LSLLKK293 pKa = 10.17TPTVSANAKK302 pKa = 7.85LTSGSSTMTAFSAYY316 pKa = 8.97IANSSQQNFNCSYY329 pKa = 11.1YY330 pKa = 9.65LQQWLVDD337 pKa = 3.8GLIMTSLYY345 pKa = 10.8LKK347 pKa = 10.26LDD349 pKa = 3.66RR350 pKa = 11.84AHH352 pKa = 6.92FSNMGSDD359 pKa = 3.31EE360 pKa = 4.2SSRR363 pKa = 11.84NAKK366 pKa = 8.79WFTFWISSTSNFNLSNISEE385 pKa = 4.26PTIEE389 pKa = 4.46PSTVQWNAFLPASNYY404 pKa = 8.38SQPASFQYY412 pKa = 7.27TTSGGVTNMYY422 pKa = 8.54YY423 pKa = 10.08QPSSGAMQTFLPVTTGDD440 pKa = 3.7WANTTYY446 pKa = 11.25DD447 pKa = 3.7PGSVNICALPVSVASSASSSRR468 pKa = 11.84MMICFNFRR476 pKa = 11.84CTNSGIFNSNATTGTMSIGPIFFTCPASAYY506 pKa = 7.25TAPP509 pKa = 4.03
MM1 pKa = 7.51SSAPFHH7 pKa = 6.34GMSISGRR14 pKa = 11.84KK15 pKa = 9.14RR16 pKa = 11.84SSNASNDD23 pKa = 2.91EE24 pKa = 4.15AEE26 pKa = 4.42PVKK29 pKa = 10.72APKK32 pKa = 9.81LQAGARR38 pKa = 11.84VTINAQNAEE47 pKa = 4.35LDD49 pKa = 3.61LVYY52 pKa = 10.23PFWYY56 pKa = 9.92EE57 pKa = 3.97STSTGGNPSVNPPLLDD73 pKa = 3.61PSGPLYY79 pKa = 10.46VQSNMLKK86 pKa = 9.83VRR88 pKa = 11.84TAAPVEE94 pKa = 4.2VANGSITLAYY104 pKa = 10.41DD105 pKa = 3.33DD106 pKa = 5.01TLTLDD111 pKa = 4.58DD112 pKa = 4.52QNKK115 pKa = 9.39LQINVEE121 pKa = 4.15PNGPLRR127 pKa = 11.84STPDD131 pKa = 3.54GLDD134 pKa = 3.33LAIDD138 pKa = 3.66STLEE142 pKa = 3.58VDD144 pKa = 3.05NWEE147 pKa = 4.85LGVKK151 pKa = 10.0LNPAGPIDD159 pKa = 4.01ASAAGLDD166 pKa = 3.6INVDD170 pKa = 3.42EE171 pKa = 4.61TLLVAEE177 pKa = 5.32DD178 pKa = 3.9STNQRR183 pKa = 11.84YY184 pKa = 8.55EE185 pKa = 4.22LGVHH189 pKa = 6.76LNPNGPITADD199 pKa = 3.23SDD201 pKa = 4.26GLDD204 pKa = 3.85LEE206 pKa = 5.04INPEE210 pKa = 4.17TLTVTNDD217 pKa = 3.11SNTGGVLSVVLKK229 pKa = 9.94PQGGLQSSLLGIGVAVDD246 pKa = 3.38RR247 pKa = 11.84TLTIDD252 pKa = 3.31QNTVEE257 pKa = 4.4VKK259 pKa = 10.08IDD261 pKa = 3.36PTGPLTSSDD270 pKa = 3.46NGLSLDD276 pKa = 3.71YY277 pKa = 9.8DD278 pKa = 3.8TTDD281 pKa = 3.83FSVQAGKK288 pKa = 10.8LSLLKK293 pKa = 10.17TPTVSANAKK302 pKa = 7.85LTSGSSTMTAFSAYY316 pKa = 8.97IANSSQQNFNCSYY329 pKa = 11.1YY330 pKa = 9.65LQQWLVDD337 pKa = 3.8GLIMTSLYY345 pKa = 10.8LKK347 pKa = 10.26LDD349 pKa = 3.66RR350 pKa = 11.84AHH352 pKa = 6.92FSNMGSDD359 pKa = 3.31EE360 pKa = 4.2SSRR363 pKa = 11.84NAKK366 pKa = 8.79WFTFWISSTSNFNLSNISEE385 pKa = 4.26PTIEE389 pKa = 4.46PSTVQWNAFLPASNYY404 pKa = 8.38SQPASFQYY412 pKa = 7.27TTSGGVTNMYY422 pKa = 8.54YY423 pKa = 10.08QPSSGAMQTFLPVTTGDD440 pKa = 3.7WANTTYY446 pKa = 11.25DD447 pKa = 3.7PGSVNICALPVSVASSASSSRR468 pKa = 11.84MMICFNFRR476 pKa = 11.84CTNSGIFNSNATTGTMSIGPIFFTCPASAYY506 pKa = 7.25TAPP509 pKa = 4.03
Molecular weight: 54.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U5NHK2|U5NHK2_9ADEN DNA-binding protein OS=Turkey aviadenovirus 4 OX=1408257 GN=DBP PE=3 SV=1
MM1 pKa = 7.68SILISPNDD9 pKa = 3.25NRR11 pKa = 11.84GWGMRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84SRR20 pKa = 11.84SSMRR24 pKa = 11.84GVGTRR29 pKa = 11.84RR30 pKa = 11.84PRR32 pKa = 11.84RR33 pKa = 11.84SRR35 pKa = 11.84MTLRR39 pKa = 11.84TLLGLGTVSRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84NRR54 pKa = 11.84RR55 pKa = 11.84TRR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84SSRR62 pKa = 11.84PASTTSRR69 pKa = 11.84LVVVRR74 pKa = 11.84TRR76 pKa = 11.84RR77 pKa = 11.84GGRR80 pKa = 3.17
MM1 pKa = 7.68SILISPNDD9 pKa = 3.25NRR11 pKa = 11.84GWGMRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84SRR20 pKa = 11.84SSMRR24 pKa = 11.84GVGTRR29 pKa = 11.84RR30 pKa = 11.84PRR32 pKa = 11.84RR33 pKa = 11.84SRR35 pKa = 11.84MTLRR39 pKa = 11.84TLLGLGTVSRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84NRR54 pKa = 11.84RR55 pKa = 11.84TRR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84SSRR62 pKa = 11.84PASTTSRR69 pKa = 11.84LVVVRR74 pKa = 11.84TRR76 pKa = 11.84RR77 pKa = 11.84GGRR80 pKa = 3.17
Molecular weight: 9.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
12730 |
67 |
1268 |
385.8 |
43.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.913 ± 0.376 | 1.948 ± 0.246 |
5.499 ± 0.224 | 5.907 ± 0.441 |
4.108 ± 0.27 | 5.947 ± 0.332 |
2.537 ± 0.313 | 4.478 ± 0.17 |
4.116 ± 0.377 | 8.578 ± 0.4 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.632 ± 0.173 | 4.588 ± 0.319 |
6.19 ± 0.288 | 3.778 ± 0.192 |
6.701 ± 0.458 | 7.714 ± 0.413 |
6.237 ± 0.364 | 6.394 ± 0.318 |
1.445 ± 0.14 | 4.289 ± 0.278 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
