
Chobar Gorge virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus
Average proteome isoelectric point is 7.37
Get precalculated fractions of proteins
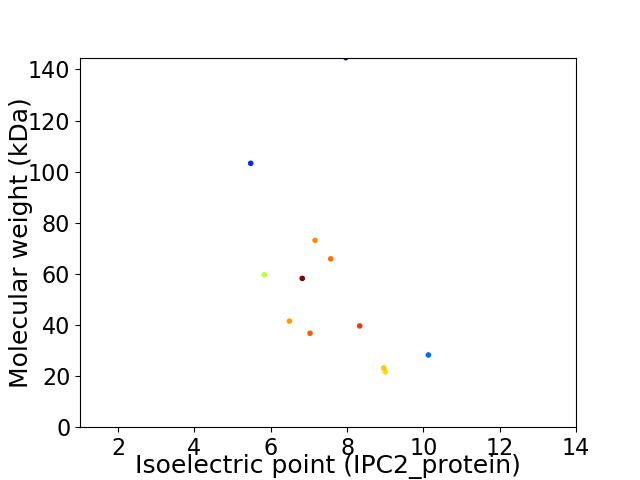
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H4M948|A0A0H4M948_9REOV Core protein VP7 OS=Chobar Gorge virus OX=1679172 GN=VP7 PE=3 SV=1
MM1 pKa = 7.77AEE3 pKa = 3.44QAAAADD9 pKa = 3.7QRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84NPDD16 pKa = 3.0PTAAPYY22 pKa = 10.72LRR24 pKa = 11.84GEE26 pKa = 4.4EE27 pKa = 4.1VRR29 pKa = 11.84DD30 pKa = 3.85DD31 pKa = 4.09NGPLLSVFALQEE43 pKa = 4.01IINKK47 pKa = 8.4VRR49 pKa = 11.84EE50 pKa = 4.16AQTRR54 pKa = 11.84LASSVAEE61 pKa = 4.38TPTASPQVEE70 pKa = 4.06AVLKK74 pKa = 10.71GISDD78 pKa = 4.3LDD80 pKa = 3.85SVKK83 pKa = 10.82GYY85 pKa = 8.6TIQNTVPISYY95 pKa = 9.33IHH97 pKa = 6.63EE98 pKa = 4.24AVQSDD103 pKa = 3.46EE104 pKa = 4.1RR105 pKa = 11.84FFRR108 pKa = 11.84IDD110 pKa = 2.7RR111 pKa = 11.84HH112 pKa = 4.7MLEE115 pKa = 4.4TSAVAADD122 pKa = 4.15VDD124 pKa = 4.14CDD126 pKa = 4.1DD127 pKa = 4.34PQGLITAIVKK137 pKa = 10.12RR138 pKa = 11.84IHH140 pKa = 6.29YY141 pKa = 9.59YY142 pKa = 9.37RR143 pKa = 11.84DD144 pKa = 3.12KK145 pKa = 11.55GSFLLWNTPTHH156 pKa = 6.53FIEE159 pKa = 4.5GRR161 pKa = 11.84EE162 pKa = 4.14VVDD165 pKa = 3.97QAALGVDD172 pKa = 3.02IDD174 pKa = 3.92SLLRR178 pKa = 11.84AVHH181 pKa = 5.44PRR183 pKa = 11.84EE184 pKa = 3.27RR185 pKa = 11.84HH186 pKa = 5.26RR187 pKa = 11.84IQRR190 pKa = 11.84EE191 pKa = 4.1LEE193 pKa = 3.67MHH195 pKa = 6.61LVMNAEE201 pKa = 4.03VRR203 pKa = 11.84GVQVDD208 pKa = 4.21TYY210 pKa = 8.85TTALPQTVYY219 pKa = 10.28EE220 pKa = 4.08VHH222 pKa = 6.34RR223 pKa = 11.84QLTEE227 pKa = 3.85YY228 pKa = 10.73VLEE231 pKa = 4.26GQRR234 pKa = 11.84GVFHH238 pKa = 7.47EE239 pKa = 4.29SMRR242 pKa = 11.84WLQMYY247 pKa = 10.82GEE249 pKa = 4.39FKK251 pKa = 10.77DD252 pKa = 4.94LEE254 pKa = 4.25FSEE257 pKa = 5.33EE258 pKa = 4.45LLTDD262 pKa = 3.65IYY264 pKa = 10.81CTDD267 pKa = 4.16TIYY270 pKa = 11.16CMSYY274 pKa = 10.05HH275 pKa = 6.7LPVNPTVIWEE285 pKa = 4.18VPRR288 pKa = 11.84CGVANLLMNAALGIPTGRR306 pKa = 11.84YY307 pKa = 8.86INPSPKK313 pKa = 9.3IASITVTSRR322 pKa = 11.84VTTTSAFSQMQSIVPTEE339 pKa = 3.61AQMNDD344 pKa = 2.55VRR346 pKa = 11.84KK347 pKa = 9.88IYY349 pKa = 10.11LALMFPNQIVIDD361 pKa = 3.99VRR363 pKa = 11.84AEE365 pKa = 3.78PGNTVDD371 pKa = 4.06QVVHH375 pKa = 6.35AVAGVVGKK383 pKa = 10.69LMFSFGPNIFNITARR398 pKa = 11.84TARR401 pKa = 11.84LLDD404 pKa = 4.07RR405 pKa = 11.84ACSMYY410 pKa = 10.74IRR412 pKa = 11.84MTTDD416 pKa = 2.76DD417 pKa = 3.86RR418 pKa = 11.84RR419 pKa = 11.84TIRR422 pKa = 11.84HH423 pKa = 5.73GRR425 pKa = 11.84TGDD428 pKa = 3.67PLDD431 pKa = 3.63FLIIQGQRR439 pKa = 11.84QVDD442 pKa = 4.17CNQLANDD449 pKa = 3.81PHH451 pKa = 6.38TGAGYY456 pKa = 9.64NAWRR460 pKa = 11.84VDD462 pKa = 3.53DD463 pKa = 5.42LGDD466 pKa = 3.63RR467 pKa = 11.84EE468 pKa = 4.41CPYY471 pKa = 8.84PHH473 pKa = 6.2VRR475 pKa = 11.84RR476 pKa = 11.84FIKK479 pKa = 10.6YY480 pKa = 10.18LGYY483 pKa = 9.08TPEE486 pKa = 6.01DD487 pKa = 3.77IIDD490 pKa = 3.57EE491 pKa = 4.36RR492 pKa = 11.84FTGQDD497 pKa = 2.68HH498 pKa = 7.22RR499 pKa = 11.84FPLMDD504 pKa = 4.34LLGNVLAVAGHH515 pKa = 4.91TNEE518 pKa = 3.97RR519 pKa = 11.84NYY521 pKa = 11.05LEE523 pKa = 5.48GMLQHH528 pKa = 6.06HH529 pKa = 6.56VVRR532 pKa = 11.84FAHH535 pKa = 7.13ISQICNRR542 pKa = 11.84DD543 pKa = 3.25LSSAFSAPDD552 pKa = 3.29DD553 pKa = 3.76RR554 pKa = 11.84FAQLGEE560 pKa = 4.07QIGNGWIAGDD570 pKa = 4.08GPLVLDD576 pKa = 4.11VSYY579 pKa = 10.72HH580 pKa = 6.39AIWFAYY586 pKa = 9.13KK587 pKa = 9.67MRR589 pKa = 11.84FLPISRR595 pKa = 11.84PDD597 pKa = 3.4MQLTQPLIEE606 pKa = 5.01SIYY609 pKa = 10.57GSQISIAKK617 pKa = 9.23YY618 pKa = 7.79QAQRR622 pKa = 11.84IDD624 pKa = 3.48DD625 pKa = 4.24FVRR628 pKa = 11.84EE629 pKa = 4.21NPEE632 pKa = 3.91SFPGIRR638 pKa = 11.84PMDD641 pKa = 3.18VWKK644 pKa = 10.69CVVSEE649 pKa = 3.97LPEE652 pKa = 4.67AIRR655 pKa = 11.84HH656 pKa = 4.47VLEE659 pKa = 5.29LVGQHH664 pKa = 6.12AFINGRR670 pKa = 11.84DD671 pKa = 3.49INLWKK676 pKa = 9.1YY677 pKa = 7.8TPAMQEE683 pKa = 4.23SLPLLCEE690 pKa = 3.71RR691 pKa = 11.84AAWDD695 pKa = 3.49AFRR698 pKa = 11.84GPEE701 pKa = 3.4AVMFTRR707 pKa = 11.84EE708 pKa = 4.07VYY710 pKa = 9.98LHH712 pKa = 6.9RR713 pKa = 11.84EE714 pKa = 4.33IIPEE718 pKa = 3.81FTLEE722 pKa = 3.95DD723 pKa = 3.56VEE725 pKa = 4.51KK726 pKa = 10.46FRR728 pKa = 11.84RR729 pKa = 11.84DD730 pKa = 2.92AEE732 pKa = 4.56YY733 pKa = 9.43YY734 pKa = 10.21TNIHH738 pKa = 6.69LNTPPADD745 pKa = 3.12EE746 pKa = 3.66RR747 pKa = 11.84VYY749 pKa = 11.19LNRR752 pKa = 11.84SSMLQRR758 pKa = 11.84AGEE761 pKa = 4.13GRR763 pKa = 11.84LNIAMRR769 pKa = 11.84RR770 pKa = 11.84YY771 pKa = 10.3LDD773 pKa = 4.17DD774 pKa = 3.77GLYY777 pKa = 9.89VQVGNILRR785 pKa = 11.84PLRR788 pKa = 11.84FQVYY792 pKa = 9.52KK793 pKa = 10.71ALPPLDD799 pKa = 3.52VRR801 pKa = 11.84EE802 pKa = 4.36ALPYY806 pKa = 9.74TYY808 pKa = 10.43TRR810 pKa = 11.84IKK812 pKa = 10.86NDD814 pKa = 2.98GPTARR819 pKa = 11.84VNVSLSKK826 pKa = 10.4SVHH829 pKa = 5.56GFLMLYY835 pKa = 9.11TADD838 pKa = 3.56EE839 pKa = 4.1QAFPDD844 pKa = 3.86EE845 pKa = 4.47MTTLVPAVCLTYY857 pKa = 10.57IKK859 pKa = 9.79TPDD862 pKa = 3.32MPFQRR867 pKa = 11.84VTIDD871 pKa = 3.36DD872 pKa = 3.84ALGVVMRR879 pKa = 11.84DD880 pKa = 2.98VMAVRR885 pKa = 11.84GKK887 pKa = 10.15IRR889 pKa = 11.84IHH891 pKa = 7.26DD892 pKa = 3.64LTAALKK898 pKa = 10.45AGSKK902 pKa = 9.71FASPQLDD909 pKa = 3.03
MM1 pKa = 7.77AEE3 pKa = 3.44QAAAADD9 pKa = 3.7QRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84NPDD16 pKa = 3.0PTAAPYY22 pKa = 10.72LRR24 pKa = 11.84GEE26 pKa = 4.4EE27 pKa = 4.1VRR29 pKa = 11.84DD30 pKa = 3.85DD31 pKa = 4.09NGPLLSVFALQEE43 pKa = 4.01IINKK47 pKa = 8.4VRR49 pKa = 11.84EE50 pKa = 4.16AQTRR54 pKa = 11.84LASSVAEE61 pKa = 4.38TPTASPQVEE70 pKa = 4.06AVLKK74 pKa = 10.71GISDD78 pKa = 4.3LDD80 pKa = 3.85SVKK83 pKa = 10.82GYY85 pKa = 8.6TIQNTVPISYY95 pKa = 9.33IHH97 pKa = 6.63EE98 pKa = 4.24AVQSDD103 pKa = 3.46EE104 pKa = 4.1RR105 pKa = 11.84FFRR108 pKa = 11.84IDD110 pKa = 2.7RR111 pKa = 11.84HH112 pKa = 4.7MLEE115 pKa = 4.4TSAVAADD122 pKa = 4.15VDD124 pKa = 4.14CDD126 pKa = 4.1DD127 pKa = 4.34PQGLITAIVKK137 pKa = 10.12RR138 pKa = 11.84IHH140 pKa = 6.29YY141 pKa = 9.59YY142 pKa = 9.37RR143 pKa = 11.84DD144 pKa = 3.12KK145 pKa = 11.55GSFLLWNTPTHH156 pKa = 6.53FIEE159 pKa = 4.5GRR161 pKa = 11.84EE162 pKa = 4.14VVDD165 pKa = 3.97QAALGVDD172 pKa = 3.02IDD174 pKa = 3.92SLLRR178 pKa = 11.84AVHH181 pKa = 5.44PRR183 pKa = 11.84EE184 pKa = 3.27RR185 pKa = 11.84HH186 pKa = 5.26RR187 pKa = 11.84IQRR190 pKa = 11.84EE191 pKa = 4.1LEE193 pKa = 3.67MHH195 pKa = 6.61LVMNAEE201 pKa = 4.03VRR203 pKa = 11.84GVQVDD208 pKa = 4.21TYY210 pKa = 8.85TTALPQTVYY219 pKa = 10.28EE220 pKa = 4.08VHH222 pKa = 6.34RR223 pKa = 11.84QLTEE227 pKa = 3.85YY228 pKa = 10.73VLEE231 pKa = 4.26GQRR234 pKa = 11.84GVFHH238 pKa = 7.47EE239 pKa = 4.29SMRR242 pKa = 11.84WLQMYY247 pKa = 10.82GEE249 pKa = 4.39FKK251 pKa = 10.77DD252 pKa = 4.94LEE254 pKa = 4.25FSEE257 pKa = 5.33EE258 pKa = 4.45LLTDD262 pKa = 3.65IYY264 pKa = 10.81CTDD267 pKa = 4.16TIYY270 pKa = 11.16CMSYY274 pKa = 10.05HH275 pKa = 6.7LPVNPTVIWEE285 pKa = 4.18VPRR288 pKa = 11.84CGVANLLMNAALGIPTGRR306 pKa = 11.84YY307 pKa = 8.86INPSPKK313 pKa = 9.3IASITVTSRR322 pKa = 11.84VTTTSAFSQMQSIVPTEE339 pKa = 3.61AQMNDD344 pKa = 2.55VRR346 pKa = 11.84KK347 pKa = 9.88IYY349 pKa = 10.11LALMFPNQIVIDD361 pKa = 3.99VRR363 pKa = 11.84AEE365 pKa = 3.78PGNTVDD371 pKa = 4.06QVVHH375 pKa = 6.35AVAGVVGKK383 pKa = 10.69LMFSFGPNIFNITARR398 pKa = 11.84TARR401 pKa = 11.84LLDD404 pKa = 4.07RR405 pKa = 11.84ACSMYY410 pKa = 10.74IRR412 pKa = 11.84MTTDD416 pKa = 2.76DD417 pKa = 3.86RR418 pKa = 11.84RR419 pKa = 11.84TIRR422 pKa = 11.84HH423 pKa = 5.73GRR425 pKa = 11.84TGDD428 pKa = 3.67PLDD431 pKa = 3.63FLIIQGQRR439 pKa = 11.84QVDD442 pKa = 4.17CNQLANDD449 pKa = 3.81PHH451 pKa = 6.38TGAGYY456 pKa = 9.64NAWRR460 pKa = 11.84VDD462 pKa = 3.53DD463 pKa = 5.42LGDD466 pKa = 3.63RR467 pKa = 11.84EE468 pKa = 4.41CPYY471 pKa = 8.84PHH473 pKa = 6.2VRR475 pKa = 11.84RR476 pKa = 11.84FIKK479 pKa = 10.6YY480 pKa = 10.18LGYY483 pKa = 9.08TPEE486 pKa = 6.01DD487 pKa = 3.77IIDD490 pKa = 3.57EE491 pKa = 4.36RR492 pKa = 11.84FTGQDD497 pKa = 2.68HH498 pKa = 7.22RR499 pKa = 11.84FPLMDD504 pKa = 4.34LLGNVLAVAGHH515 pKa = 4.91TNEE518 pKa = 3.97RR519 pKa = 11.84NYY521 pKa = 11.05LEE523 pKa = 5.48GMLQHH528 pKa = 6.06HH529 pKa = 6.56VVRR532 pKa = 11.84FAHH535 pKa = 7.13ISQICNRR542 pKa = 11.84DD543 pKa = 3.25LSSAFSAPDD552 pKa = 3.29DD553 pKa = 3.76RR554 pKa = 11.84FAQLGEE560 pKa = 4.07QIGNGWIAGDD570 pKa = 4.08GPLVLDD576 pKa = 4.11VSYY579 pKa = 10.72HH580 pKa = 6.39AIWFAYY586 pKa = 9.13KK587 pKa = 9.67MRR589 pKa = 11.84FLPISRR595 pKa = 11.84PDD597 pKa = 3.4MQLTQPLIEE606 pKa = 5.01SIYY609 pKa = 10.57GSQISIAKK617 pKa = 9.23YY618 pKa = 7.79QAQRR622 pKa = 11.84IDD624 pKa = 3.48DD625 pKa = 4.24FVRR628 pKa = 11.84EE629 pKa = 4.21NPEE632 pKa = 3.91SFPGIRR638 pKa = 11.84PMDD641 pKa = 3.18VWKK644 pKa = 10.69CVVSEE649 pKa = 3.97LPEE652 pKa = 4.67AIRR655 pKa = 11.84HH656 pKa = 4.47VLEE659 pKa = 5.29LVGQHH664 pKa = 6.12AFINGRR670 pKa = 11.84DD671 pKa = 3.49INLWKK676 pKa = 9.1YY677 pKa = 7.8TPAMQEE683 pKa = 4.23SLPLLCEE690 pKa = 3.71RR691 pKa = 11.84AAWDD695 pKa = 3.49AFRR698 pKa = 11.84GPEE701 pKa = 3.4AVMFTRR707 pKa = 11.84EE708 pKa = 4.07VYY710 pKa = 9.98LHH712 pKa = 6.9RR713 pKa = 11.84EE714 pKa = 4.33IIPEE718 pKa = 3.81FTLEE722 pKa = 3.95DD723 pKa = 3.56VEE725 pKa = 4.51KK726 pKa = 10.46FRR728 pKa = 11.84RR729 pKa = 11.84DD730 pKa = 2.92AEE732 pKa = 4.56YY733 pKa = 9.43YY734 pKa = 10.21TNIHH738 pKa = 6.69LNTPPADD745 pKa = 3.12EE746 pKa = 3.66RR747 pKa = 11.84VYY749 pKa = 11.19LNRR752 pKa = 11.84SSMLQRR758 pKa = 11.84AGEE761 pKa = 4.13GRR763 pKa = 11.84LNIAMRR769 pKa = 11.84RR770 pKa = 11.84YY771 pKa = 10.3LDD773 pKa = 4.17DD774 pKa = 3.77GLYY777 pKa = 9.89VQVGNILRR785 pKa = 11.84PLRR788 pKa = 11.84FQVYY792 pKa = 9.52KK793 pKa = 10.71ALPPLDD799 pKa = 3.52VRR801 pKa = 11.84EE802 pKa = 4.36ALPYY806 pKa = 9.74TYY808 pKa = 10.43TRR810 pKa = 11.84IKK812 pKa = 10.86NDD814 pKa = 2.98GPTARR819 pKa = 11.84VNVSLSKK826 pKa = 10.4SVHH829 pKa = 5.56GFLMLYY835 pKa = 9.11TADD838 pKa = 3.56EE839 pKa = 4.1QAFPDD844 pKa = 3.86EE845 pKa = 4.47MTTLVPAVCLTYY857 pKa = 10.57IKK859 pKa = 9.79TPDD862 pKa = 3.32MPFQRR867 pKa = 11.84VTIDD871 pKa = 3.36DD872 pKa = 3.84ALGVVMRR879 pKa = 11.84DD880 pKa = 2.98VMAVRR885 pKa = 11.84GKK887 pKa = 10.15IRR889 pKa = 11.84IHH891 pKa = 7.26DD892 pKa = 3.64LTAALKK898 pKa = 10.45AGSKK902 pKa = 9.71FASPQLDD909 pKa = 3.03
Molecular weight: 103.27 kDa
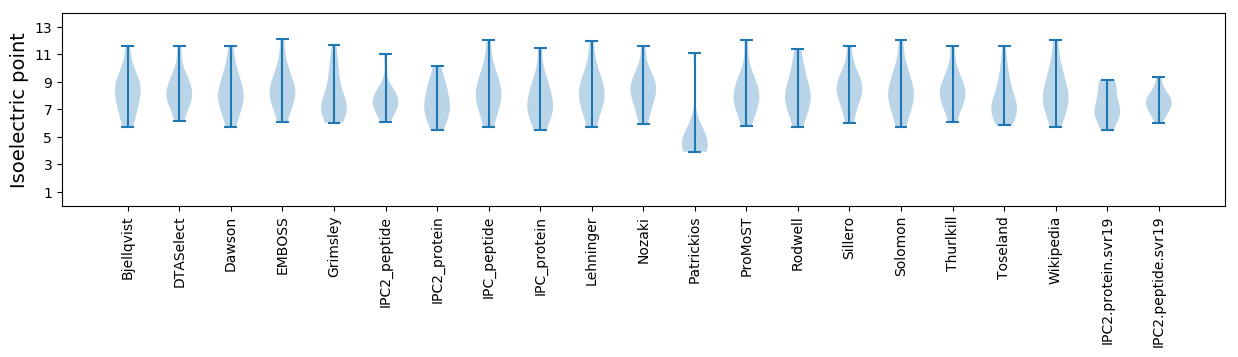
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H4M5E8|A0A0H4M5E8_9REOV Core protein VP4 OS=Chobar Gorge virus OX=1679172 GN=VP3 PE=3 SV=1
MM1 pKa = 7.51SSEE4 pKa = 4.14RR5 pKa = 11.84VKK7 pKa = 11.13LNSPSGSSMSGLLLGKK23 pKa = 9.79KK24 pKa = 5.91WWKK27 pKa = 10.09EE28 pKa = 3.21IRR30 pKa = 11.84KK31 pKa = 8.97LLRR34 pKa = 11.84ISARR38 pKa = 11.84KK39 pKa = 9.74EE40 pKa = 3.55EE41 pKa = 4.45MLTGEE46 pKa = 4.66YY47 pKa = 10.19DD48 pKa = 3.12RR49 pKa = 11.84KK50 pKa = 8.95TYY52 pKa = 11.22AEE54 pKa = 4.24TLSALRR60 pKa = 11.84TQYY63 pKa = 8.89GTQILKK69 pKa = 10.45RR70 pKa = 11.84EE71 pKa = 4.06TRR73 pKa = 11.84MQMMLNSMPVAQLARR88 pKa = 11.84LPTPTRR94 pKa = 11.84RR95 pKa = 11.84LQRR98 pKa = 11.84TVGTVIVEE106 pKa = 4.12LQRR109 pKa = 11.84TINQTGLEE117 pKa = 4.02RR118 pKa = 11.84WRR120 pKa = 11.84VAVRR124 pKa = 11.84YY125 pKa = 9.28ALTRR129 pKa = 11.84IPRR132 pKa = 11.84AWRR135 pKa = 11.84RR136 pKa = 11.84SEE138 pKa = 4.16RR139 pKa = 11.84IRR141 pKa = 11.84AARR144 pKa = 11.84LFAYY148 pKa = 10.18LRR150 pKa = 11.84AEE152 pKa = 4.22TEE154 pKa = 3.88EE155 pKa = 3.88QRR157 pKa = 11.84QVLATLTRR165 pKa = 11.84LQVEE169 pKa = 4.36EE170 pKa = 4.64RR171 pKa = 11.84LLSTSQISEE180 pKa = 4.46FVTDD184 pKa = 3.35WQEE187 pKa = 3.6KK188 pKa = 8.92LRR190 pKa = 11.84QIFGFFPHH198 pKa = 6.3PQKK201 pKa = 10.94LIRR204 pKa = 11.84QLYY207 pKa = 7.59SSFWVRR213 pKa = 11.84RR214 pKa = 11.84SLSRR218 pKa = 11.84SEE220 pKa = 3.79SRR222 pKa = 11.84RR223 pKa = 11.84IRR225 pKa = 11.84SLNRR229 pKa = 11.84KK230 pKa = 6.64MRR232 pKa = 11.84RR233 pKa = 11.84LGLEE237 pKa = 3.93GG238 pKa = 3.57
MM1 pKa = 7.51SSEE4 pKa = 4.14RR5 pKa = 11.84VKK7 pKa = 11.13LNSPSGSSMSGLLLGKK23 pKa = 9.79KK24 pKa = 5.91WWKK27 pKa = 10.09EE28 pKa = 3.21IRR30 pKa = 11.84KK31 pKa = 8.97LLRR34 pKa = 11.84ISARR38 pKa = 11.84KK39 pKa = 9.74EE40 pKa = 3.55EE41 pKa = 4.45MLTGEE46 pKa = 4.66YY47 pKa = 10.19DD48 pKa = 3.12RR49 pKa = 11.84KK50 pKa = 8.95TYY52 pKa = 11.22AEE54 pKa = 4.24TLSALRR60 pKa = 11.84TQYY63 pKa = 8.89GTQILKK69 pKa = 10.45RR70 pKa = 11.84EE71 pKa = 4.06TRR73 pKa = 11.84MQMMLNSMPVAQLARR88 pKa = 11.84LPTPTRR94 pKa = 11.84RR95 pKa = 11.84LQRR98 pKa = 11.84TVGTVIVEE106 pKa = 4.12LQRR109 pKa = 11.84TINQTGLEE117 pKa = 4.02RR118 pKa = 11.84WRR120 pKa = 11.84VAVRR124 pKa = 11.84YY125 pKa = 9.28ALTRR129 pKa = 11.84IPRR132 pKa = 11.84AWRR135 pKa = 11.84RR136 pKa = 11.84SEE138 pKa = 4.16RR139 pKa = 11.84IRR141 pKa = 11.84AARR144 pKa = 11.84LFAYY148 pKa = 10.18LRR150 pKa = 11.84AEE152 pKa = 4.22TEE154 pKa = 3.88EE155 pKa = 3.88QRR157 pKa = 11.84QVLATLTRR165 pKa = 11.84LQVEE169 pKa = 4.36EE170 pKa = 4.64RR171 pKa = 11.84LLSTSQISEE180 pKa = 4.46FVTDD184 pKa = 3.35WQEE187 pKa = 3.6KK188 pKa = 8.92LRR190 pKa = 11.84QIFGFFPHH198 pKa = 6.3PQKK201 pKa = 10.94LIRR204 pKa = 11.84QLYY207 pKa = 7.59SSFWVRR213 pKa = 11.84RR214 pKa = 11.84SLSRR218 pKa = 11.84SEE220 pKa = 3.79SRR222 pKa = 11.84RR223 pKa = 11.84IRR225 pKa = 11.84SLNRR229 pKa = 11.84KK230 pKa = 6.64MRR232 pKa = 11.84RR233 pKa = 11.84LGLEE237 pKa = 3.93GG238 pKa = 3.57
Molecular weight: 28.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6180 |
192 |
1284 |
515.0 |
57.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.078 ± 0.71 | 1.343 ± 0.224 |
5.502 ± 0.439 | 6.068 ± 0.347 |
3.058 ± 0.299 | 5.906 ± 0.225 |
2.476 ± 0.265 | 5.55 ± 0.244 |
3.333 ± 0.426 | 9.32 ± 0.667 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.816 ± 0.18 | 2.783 ± 0.298 |
4.968 ± 0.392 | 3.932 ± 0.232 |
8.948 ± 0.459 | 6.246 ± 0.463 |
6.634 ± 0.351 | 7.201 ± 0.323 |
1.262 ± 0.292 | 3.576 ± 0.26 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
