
Breoghania corrubedonensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Breoghaniaceae; Breoghania
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
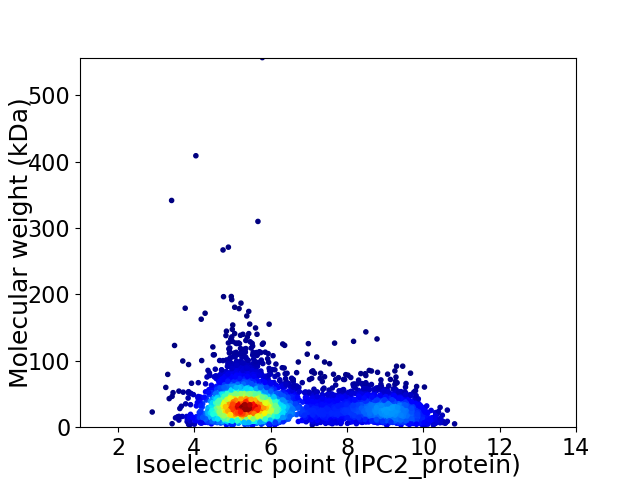
Virtual 2D-PAGE plot for 4681 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T5VGY0|A0A2T5VGY0_9RHIZ Transcription termination/antitermination protein NusA OS=Breoghania corrubedonensis OX=665038 GN=nusA PE=3 SV=1
MM1 pKa = 7.47IGAKK5 pKa = 9.4RR6 pKa = 11.84VLAAIGLLLAATIATAHH23 pKa = 6.15TADD26 pKa = 3.65AQPTFSAQFSPSTIAAGAEE45 pKa = 3.79SRR47 pKa = 11.84LIFTITNGSGSPVTAMTFTDD67 pKa = 3.97TLPAGTTVAASPRR80 pKa = 11.84GEE82 pKa = 4.26TTCIDD87 pKa = 3.36GVVTVNASDD96 pKa = 3.46TFTFEE101 pKa = 4.44GNGGASGASVAADD114 pKa = 3.52SSCTVSIYY122 pKa = 9.3VTASTGGAYY131 pKa = 10.77NNTSGDD137 pKa = 3.84LTSSAGNSGTASATLTVDD155 pKa = 3.81AARR158 pKa = 11.84LEE160 pKa = 4.07FDD162 pKa = 3.73MAFSPATVGIGAASRR177 pKa = 11.84ITYY180 pKa = 8.0TITKK184 pKa = 8.58PNSGVVTGVAFNNTLPTGLQVASPSSVATSCTGGAASATTGSQTVQLSGAFFTAAGSCTVTVDD247 pKa = 3.39VTNTGAGTYY256 pKa = 10.22VNSAEE261 pKa = 4.38LSSSAGTSGAATATLTVTAPTGDD284 pKa = 2.92ITLIKK289 pKa = 10.41QFARR293 pKa = 11.84TAAAPGGTIEE303 pKa = 4.14MTYY306 pKa = 10.03TLANRR311 pKa = 11.84SSSSAATAITFTDD324 pKa = 4.18DD325 pKa = 3.99LDD327 pKa = 4.16AALSGLTASALPATGFCGVGSQLTGSSTLTLSGATLGSRR366 pKa = 11.84EE367 pKa = 4.27SCSFTVTLDD376 pKa = 3.58VPASASDD383 pKa = 3.5TTVTSTTSSISGQLGGSPVTGDD405 pKa = 3.42PASASFNVVTTTPVMTKK422 pKa = 10.17SFTDD426 pKa = 3.71DD427 pKa = 3.42PVSAGGTVTLQYY439 pKa = 10.81VISNPSTITALGSIAFTDD457 pKa = 3.77SLAAMADD464 pKa = 3.48AAVTAGTGSNLCGSGSFLFTQEE486 pKa = 3.55ISGTPSVVLNSGTLAAGASCTLTLTLTLPANTPAGDD522 pKa = 3.88YY523 pKa = 10.88ASTSSDD529 pKa = 3.33VTSDD533 pKa = 3.12GGTGAPASDD542 pKa = 4.2TLQVSPPDD550 pKa = 3.6INVSLSKK557 pKa = 9.86TFTDD561 pKa = 3.94DD562 pKa = 3.21PVRR565 pKa = 11.84PGDD568 pKa = 3.67TVTLEE573 pKa = 3.71FTLRR577 pKa = 11.84NDD579 pKa = 4.4DD580 pKa = 3.74EE581 pKa = 4.48ATAITDD587 pKa = 4.38LAFTDD592 pKa = 5.23DD593 pKa = 4.54LDD595 pKa = 5.56AMLTGAVATGLPASNVCGTGSTVSGTSTIALSGGNLAASDD635 pKa = 3.88EE636 pKa = 4.78CTFSVTVQVPGGAAAGSYY654 pKa = 10.73PNTTSDD660 pKa = 3.16VSGDD664 pKa = 3.6SGSGIMTATGSAATGTLVVSDD685 pKa = 3.9VLAVTGTMSFTDD697 pKa = 3.96DD698 pKa = 3.73PVVAGSVVTLQYY710 pKa = 11.35VLSNPNTADD719 pKa = 3.66DD720 pKa = 3.77ATSISFSDD728 pKa = 3.6NLNGLGLPSPGLVLSGSLPSDD749 pKa = 3.25PCGTGSSLSLSSGGTAGTLSLSGGNILASGSCTFSIQFTVPSNAAANTYY798 pKa = 11.06GNTTSVISSTVNSTPLTSASGMNDD822 pKa = 3.17TLSVSSPYY830 pKa = 10.97EE831 pKa = 3.65NIIFTKK837 pKa = 10.42TFTDD841 pKa = 4.17DD842 pKa = 3.43PAEE845 pKa = 4.47AGSSVTLRR853 pKa = 11.84FVLDD857 pKa = 3.43NSVNSVAIDD866 pKa = 3.53GATFTDD872 pKa = 4.77DD873 pKa = 3.17LTAMGFAATVTADD886 pKa = 3.33SGSCSGGGSWNVTGTTTISASVSNLPGGATCTFDD920 pKa = 3.32VAVAIPSGTSPGISTNTTSTVAVTIGAGSGSVAAASDD957 pKa = 4.21DD958 pKa = 3.83LTISPTNVTLQKK970 pKa = 10.49SFSGDD975 pKa = 3.17PVLPGGTFSVTYY987 pKa = 9.42TILSPASGSALSDD1000 pKa = 2.84IRR1002 pKa = 11.84FTDD1005 pKa = 4.16DD1006 pKa = 2.84YY1007 pKa = 11.68DD1008 pKa = 3.94AVLSGLVVSGALPSNPCGAGSALAGTTSIEE1038 pKa = 4.12LTGGSLNPSEE1048 pKa = 4.42TCQFTVNLAAPSNATPNTYY1067 pKa = 9.98TSTSGALFEE1076 pKa = 4.45TAAQIAPPASDD1087 pKa = 3.19TFEE1090 pKa = 4.25VANPADD1096 pKa = 3.61NVTFALQFTDD1106 pKa = 5.26DD1107 pKa = 3.84PVAPGGTATAQFTITNPDD1125 pKa = 3.13SVSTVSGLAFTMPVDD1140 pKa = 3.89AVITGATVSSATPAAPCGGAVVSGTSSLSVSGGSLAAGANCSFTVSVLVPLGTTSGSNSFSTSTLAATANSTATTVSAASDD1221 pKa = 3.61TLVVGDD1227 pKa = 4.68APQFTVTSGDD1237 pKa = 3.67FNTSGAVGGPFNATMDD1253 pKa = 3.94YY1254 pKa = 10.68VISNTGSFPLSYY1266 pKa = 9.69TAAASQAFIGLSSTGGTIAAGGSQTVTVSITATANGFAASATPYY1310 pKa = 10.34SGTVTFTNISASGPPTEE1327 pKa = 4.3IRR1329 pKa = 11.84NVNLTVQQLGSVTVMVNSSQGDD1351 pKa = 3.74GTFTFSSSATALNGLAITTSNGAGSSAAVDD1381 pKa = 3.97VVAGSYY1387 pKa = 8.77TLSLPKK1393 pKa = 10.43AGMPDD1398 pKa = 3.44GFGLAGISCTEE1409 pKa = 3.94TGGTSNSSASVGSATATIVVEE1430 pKa = 3.98TGEE1433 pKa = 5.15AITCVFEE1440 pKa = 4.22TANSRR1445 pKa = 11.84EE1446 pKa = 3.95RR1447 pKa = 11.84TTAIINRR1454 pKa = 11.84FLSKK1458 pKa = 10.43RR1459 pKa = 11.84VDD1461 pKa = 4.41LILSNEE1467 pKa = 3.95PGQNRR1472 pKa = 11.84RR1473 pKa = 11.84IDD1475 pKa = 3.42RR1476 pKa = 11.84LRR1478 pKa = 11.84NRR1480 pKa = 11.84FGGGASGTGTPFDD1493 pKa = 3.69VTATQAGAGGVAMAFEE1509 pKa = 4.44TSLAQIRR1516 pKa = 11.84SAYY1519 pKa = 8.74VAQDD1523 pKa = 3.28RR1524 pKa = 11.84AKK1526 pKa = 10.23QALLATSGVFAHH1538 pKa = 6.43NPSGLPPEE1546 pKa = 4.09QTLWDD1551 pKa = 3.33VWFRR1555 pKa = 11.84GSFTWFEE1562 pKa = 4.64DD1563 pKa = 3.49NSDD1566 pKa = 3.83GSDD1569 pKa = 3.38SDD1571 pKa = 4.02GVFAIFHH1578 pKa = 6.77AGADD1582 pKa = 3.79YY1583 pKa = 10.87LVSDD1587 pKa = 5.44RR1588 pKa = 11.84ILVGALVQLDD1598 pKa = 3.95YY1599 pKa = 10.68MDD1601 pKa = 4.93QDD1603 pKa = 3.82FTTLNANANGVGWMAGPYY1621 pKa = 8.16ATLRR1625 pKa = 11.84LTNNLYY1631 pKa = 10.64FDD1633 pKa = 4.04TRR1635 pKa = 11.84AAWGTSFNQVSPFNTYY1651 pKa = 9.93TDD1653 pKa = 3.98DD1654 pKa = 4.13FSTTRR1659 pKa = 11.84WLVRR1663 pKa = 11.84AGLIGDD1669 pKa = 3.78WAFGAWNFRR1678 pKa = 11.84PSANVAYY1685 pKa = 9.03MQEE1688 pKa = 4.12TQEE1691 pKa = 4.78AYY1693 pKa = 9.34TDD1695 pKa = 3.86SLSVEE1700 pKa = 4.07IPEE1703 pKa = 4.01QTVGVGQLDD1712 pKa = 4.24FGPEE1716 pKa = 3.27ISYY1719 pKa = 8.75TYY1721 pKa = 8.61QTEE1724 pKa = 4.07TGYY1727 pKa = 11.22LIAPSAKK1734 pKa = 9.3LTGIWNFKK1742 pKa = 9.66RR1743 pKa = 11.84DD1744 pKa = 3.78TYY1746 pKa = 11.63GVTTTTSSAGVEE1758 pKa = 3.99MRR1760 pKa = 11.84AKK1762 pKa = 10.73AEE1764 pKa = 3.97LGLNVRR1770 pKa = 11.84MPSGVRR1776 pKa = 11.84VEE1778 pKa = 5.07ANGSYY1783 pKa = 10.75DD1784 pKa = 3.68GIGAEE1789 pKa = 5.07DD1790 pKa = 3.93YY1791 pKa = 10.89NAVSGQVKK1799 pKa = 10.31LSVPLNN1805 pKa = 3.64
MM1 pKa = 7.47IGAKK5 pKa = 9.4RR6 pKa = 11.84VLAAIGLLLAATIATAHH23 pKa = 6.15TADD26 pKa = 3.65AQPTFSAQFSPSTIAAGAEE45 pKa = 3.79SRR47 pKa = 11.84LIFTITNGSGSPVTAMTFTDD67 pKa = 3.97TLPAGTTVAASPRR80 pKa = 11.84GEE82 pKa = 4.26TTCIDD87 pKa = 3.36GVVTVNASDD96 pKa = 3.46TFTFEE101 pKa = 4.44GNGGASGASVAADD114 pKa = 3.52SSCTVSIYY122 pKa = 9.3VTASTGGAYY131 pKa = 10.77NNTSGDD137 pKa = 3.84LTSSAGNSGTASATLTVDD155 pKa = 3.81AARR158 pKa = 11.84LEE160 pKa = 4.07FDD162 pKa = 3.73MAFSPATVGIGAASRR177 pKa = 11.84ITYY180 pKa = 8.0TITKK184 pKa = 8.58PNSGVVTGVAFNNTLPTGLQVASPSSVATSCTGGAASATTGSQTVQLSGAFFTAAGSCTVTVDD247 pKa = 3.39VTNTGAGTYY256 pKa = 10.22VNSAEE261 pKa = 4.38LSSSAGTSGAATATLTVTAPTGDD284 pKa = 2.92ITLIKK289 pKa = 10.41QFARR293 pKa = 11.84TAAAPGGTIEE303 pKa = 4.14MTYY306 pKa = 10.03TLANRR311 pKa = 11.84SSSSAATAITFTDD324 pKa = 4.18DD325 pKa = 3.99LDD327 pKa = 4.16AALSGLTASALPATGFCGVGSQLTGSSTLTLSGATLGSRR366 pKa = 11.84EE367 pKa = 4.27SCSFTVTLDD376 pKa = 3.58VPASASDD383 pKa = 3.5TTVTSTTSSISGQLGGSPVTGDD405 pKa = 3.42PASASFNVVTTTPVMTKK422 pKa = 10.17SFTDD426 pKa = 3.71DD427 pKa = 3.42PVSAGGTVTLQYY439 pKa = 10.81VISNPSTITALGSIAFTDD457 pKa = 3.77SLAAMADD464 pKa = 3.48AAVTAGTGSNLCGSGSFLFTQEE486 pKa = 3.55ISGTPSVVLNSGTLAAGASCTLTLTLTLPANTPAGDD522 pKa = 3.88YY523 pKa = 10.88ASTSSDD529 pKa = 3.33VTSDD533 pKa = 3.12GGTGAPASDD542 pKa = 4.2TLQVSPPDD550 pKa = 3.6INVSLSKK557 pKa = 9.86TFTDD561 pKa = 3.94DD562 pKa = 3.21PVRR565 pKa = 11.84PGDD568 pKa = 3.67TVTLEE573 pKa = 3.71FTLRR577 pKa = 11.84NDD579 pKa = 4.4DD580 pKa = 3.74EE581 pKa = 4.48ATAITDD587 pKa = 4.38LAFTDD592 pKa = 5.23DD593 pKa = 4.54LDD595 pKa = 5.56AMLTGAVATGLPASNVCGTGSTVSGTSTIALSGGNLAASDD635 pKa = 3.88EE636 pKa = 4.78CTFSVTVQVPGGAAAGSYY654 pKa = 10.73PNTTSDD660 pKa = 3.16VSGDD664 pKa = 3.6SGSGIMTATGSAATGTLVVSDD685 pKa = 3.9VLAVTGTMSFTDD697 pKa = 3.96DD698 pKa = 3.73PVVAGSVVTLQYY710 pKa = 11.35VLSNPNTADD719 pKa = 3.66DD720 pKa = 3.77ATSISFSDD728 pKa = 3.6NLNGLGLPSPGLVLSGSLPSDD749 pKa = 3.25PCGTGSSLSLSSGGTAGTLSLSGGNILASGSCTFSIQFTVPSNAAANTYY798 pKa = 11.06GNTTSVISSTVNSTPLTSASGMNDD822 pKa = 3.17TLSVSSPYY830 pKa = 10.97EE831 pKa = 3.65NIIFTKK837 pKa = 10.42TFTDD841 pKa = 4.17DD842 pKa = 3.43PAEE845 pKa = 4.47AGSSVTLRR853 pKa = 11.84FVLDD857 pKa = 3.43NSVNSVAIDD866 pKa = 3.53GATFTDD872 pKa = 4.77DD873 pKa = 3.17LTAMGFAATVTADD886 pKa = 3.33SGSCSGGGSWNVTGTTTISASVSNLPGGATCTFDD920 pKa = 3.32VAVAIPSGTSPGISTNTTSTVAVTIGAGSGSVAAASDD957 pKa = 4.21DD958 pKa = 3.83LTISPTNVTLQKK970 pKa = 10.49SFSGDD975 pKa = 3.17PVLPGGTFSVTYY987 pKa = 9.42TILSPASGSALSDD1000 pKa = 2.84IRR1002 pKa = 11.84FTDD1005 pKa = 4.16DD1006 pKa = 2.84YY1007 pKa = 11.68DD1008 pKa = 3.94AVLSGLVVSGALPSNPCGAGSALAGTTSIEE1038 pKa = 4.12LTGGSLNPSEE1048 pKa = 4.42TCQFTVNLAAPSNATPNTYY1067 pKa = 9.98TSTSGALFEE1076 pKa = 4.45TAAQIAPPASDD1087 pKa = 3.19TFEE1090 pKa = 4.25VANPADD1096 pKa = 3.61NVTFALQFTDD1106 pKa = 5.26DD1107 pKa = 3.84PVAPGGTATAQFTITNPDD1125 pKa = 3.13SVSTVSGLAFTMPVDD1140 pKa = 3.89AVITGATVSSATPAAPCGGAVVSGTSSLSVSGGSLAAGANCSFTVSVLVPLGTTSGSNSFSTSTLAATANSTATTVSAASDD1221 pKa = 3.61TLVVGDD1227 pKa = 4.68APQFTVTSGDD1237 pKa = 3.67FNTSGAVGGPFNATMDD1253 pKa = 3.94YY1254 pKa = 10.68VISNTGSFPLSYY1266 pKa = 9.69TAAASQAFIGLSSTGGTIAAGGSQTVTVSITATANGFAASATPYY1310 pKa = 10.34SGTVTFTNISASGPPTEE1327 pKa = 4.3IRR1329 pKa = 11.84NVNLTVQQLGSVTVMVNSSQGDD1351 pKa = 3.74GTFTFSSSATALNGLAITTSNGAGSSAAVDD1381 pKa = 3.97VVAGSYY1387 pKa = 8.77TLSLPKK1393 pKa = 10.43AGMPDD1398 pKa = 3.44GFGLAGISCTEE1409 pKa = 3.94TGGTSNSSASVGSATATIVVEE1430 pKa = 3.98TGEE1433 pKa = 5.15AITCVFEE1440 pKa = 4.22TANSRR1445 pKa = 11.84EE1446 pKa = 3.95RR1447 pKa = 11.84TTAIINRR1454 pKa = 11.84FLSKK1458 pKa = 10.43RR1459 pKa = 11.84VDD1461 pKa = 4.41LILSNEE1467 pKa = 3.95PGQNRR1472 pKa = 11.84RR1473 pKa = 11.84IDD1475 pKa = 3.42RR1476 pKa = 11.84LRR1478 pKa = 11.84NRR1480 pKa = 11.84FGGGASGTGTPFDD1493 pKa = 3.69VTATQAGAGGVAMAFEE1509 pKa = 4.44TSLAQIRR1516 pKa = 11.84SAYY1519 pKa = 8.74VAQDD1523 pKa = 3.28RR1524 pKa = 11.84AKK1526 pKa = 10.23QALLATSGVFAHH1538 pKa = 6.43NPSGLPPEE1546 pKa = 4.09QTLWDD1551 pKa = 3.33VWFRR1555 pKa = 11.84GSFTWFEE1562 pKa = 4.64DD1563 pKa = 3.49NSDD1566 pKa = 3.83GSDD1569 pKa = 3.38SDD1571 pKa = 4.02GVFAIFHH1578 pKa = 6.77AGADD1582 pKa = 3.79YY1583 pKa = 10.87LVSDD1587 pKa = 5.44RR1588 pKa = 11.84ILVGALVQLDD1598 pKa = 3.95YY1599 pKa = 10.68MDD1601 pKa = 4.93QDD1603 pKa = 3.82FTTLNANANGVGWMAGPYY1621 pKa = 8.16ATLRR1625 pKa = 11.84LTNNLYY1631 pKa = 10.64FDD1633 pKa = 4.04TRR1635 pKa = 11.84AAWGTSFNQVSPFNTYY1651 pKa = 9.93TDD1653 pKa = 3.98DD1654 pKa = 4.13FSTTRR1659 pKa = 11.84WLVRR1663 pKa = 11.84AGLIGDD1669 pKa = 3.78WAFGAWNFRR1678 pKa = 11.84PSANVAYY1685 pKa = 9.03MQEE1688 pKa = 4.12TQEE1691 pKa = 4.78AYY1693 pKa = 9.34TDD1695 pKa = 3.86SLSVEE1700 pKa = 4.07IPEE1703 pKa = 4.01QTVGVGQLDD1712 pKa = 4.24FGPEE1716 pKa = 3.27ISYY1719 pKa = 8.75TYY1721 pKa = 8.61QTEE1724 pKa = 4.07TGYY1727 pKa = 11.22LIAPSAKK1734 pKa = 9.3LTGIWNFKK1742 pKa = 9.66RR1743 pKa = 11.84DD1744 pKa = 3.78TYY1746 pKa = 11.63GVTTTTSSAGVEE1758 pKa = 3.99MRR1760 pKa = 11.84AKK1762 pKa = 10.73AEE1764 pKa = 3.97LGLNVRR1770 pKa = 11.84MPSGVRR1776 pKa = 11.84VEE1778 pKa = 5.07ANGSYY1783 pKa = 10.75DD1784 pKa = 3.68GIGAEE1789 pKa = 5.07DD1790 pKa = 3.93YY1791 pKa = 10.89NAVSGQVKK1799 pKa = 10.31LSVPLNN1805 pKa = 3.64
Molecular weight: 179.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T5VF07|A0A2T5VF07_9RHIZ Dolichyl-phosphate-mannose-protein mannosyltransferase OS=Breoghania corrubedonensis OX=665038 GN=C8N35_101369 PE=4 SV=1
MM1 pKa = 6.85QRR3 pKa = 11.84LRR5 pKa = 11.84AGGNIAAGLAFVALAALFPATAFAACKK32 pKa = 9.64PPEE35 pKa = 4.45KK36 pKa = 9.66PAPPGCRR43 pKa = 11.84IKK45 pKa = 11.33GNINSDD51 pKa = 3.36GARR54 pKa = 11.84IYY56 pKa = 10.33HH57 pKa = 5.76VPGKK61 pKa = 9.86SRR63 pKa = 11.84WYY65 pKa = 10.71SRR67 pKa = 11.84TCINRR72 pKa = 11.84PGEE75 pKa = 3.77RR76 pKa = 11.84WFCSEE81 pKa = 4.01EE82 pKa = 3.79EE83 pKa = 3.83ARR85 pKa = 11.84AAGWRR90 pKa = 11.84APRR93 pKa = 11.84RR94 pKa = 3.71
MM1 pKa = 6.85QRR3 pKa = 11.84LRR5 pKa = 11.84AGGNIAAGLAFVALAALFPATAFAACKK32 pKa = 9.64PPEE35 pKa = 4.45KK36 pKa = 9.66PAPPGCRR43 pKa = 11.84IKK45 pKa = 11.33GNINSDD51 pKa = 3.36GARR54 pKa = 11.84IYY56 pKa = 10.33HH57 pKa = 5.76VPGKK61 pKa = 9.86SRR63 pKa = 11.84WYY65 pKa = 10.71SRR67 pKa = 11.84TCINRR72 pKa = 11.84PGEE75 pKa = 3.77RR76 pKa = 11.84WFCSEE81 pKa = 4.01EE82 pKa = 3.79EE83 pKa = 3.83ARR85 pKa = 11.84AAGWRR90 pKa = 11.84APRR93 pKa = 11.84RR94 pKa = 3.71
Molecular weight: 10.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1532312 |
25 |
5123 |
327.3 |
35.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.466 ± 0.048 | 0.883 ± 0.013 |
5.937 ± 0.034 | 5.998 ± 0.039 |
3.716 ± 0.022 | 8.67 ± 0.042 |
2.005 ± 0.016 | 5.293 ± 0.028 |
3.289 ± 0.029 | 9.85 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.563 ± 0.019 | 2.577 ± 0.023 |
4.957 ± 0.025 | 2.858 ± 0.02 |
7.041 ± 0.041 | 5.468 ± 0.032 |
5.482 ± 0.044 | 7.498 ± 0.031 |
1.236 ± 0.015 | 2.213 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
