
Bovine rotavirus C
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Rotavirus; Rotavirus C
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
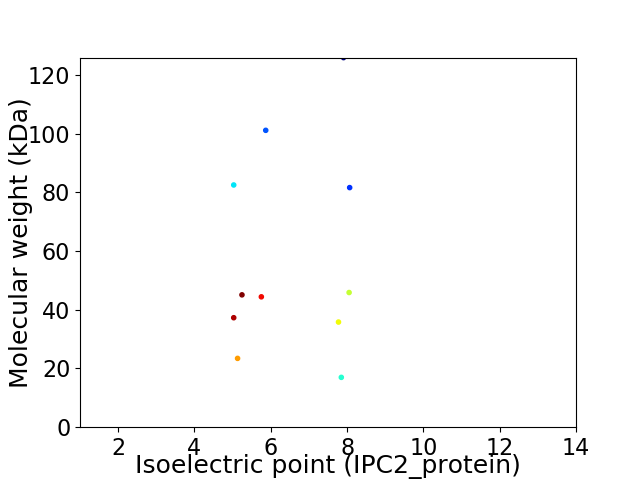
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
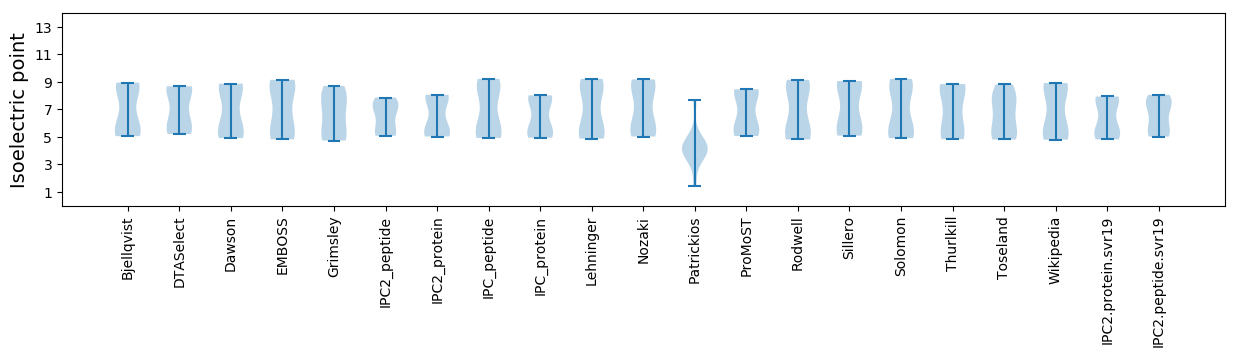
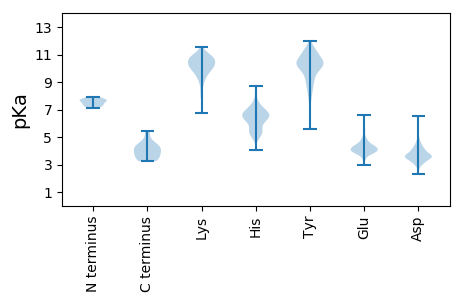
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A060NEB5|A0A060NEB5_9REOV Non-structural protein 5 OS=Bovine rotavirus C OX=31588 GN=NSP5 PE=3 SV=1
MM1 pKa = 7.33VCTTLYY7 pKa = 8.57TVCVVLCILLMYY19 pKa = 10.38ILLFRR24 pKa = 11.84KK25 pKa = 9.74VIHH28 pKa = 6.25LLIDD32 pKa = 3.23MSLIAFVISSCIGLSNAQFFANDD55 pKa = 3.36MLYY58 pKa = 10.69DD59 pKa = 3.91GNVEE63 pKa = 4.23GVINTTNIFNVEE75 pKa = 4.08SLCIYY80 pKa = 9.69FPNSAVGQPGPGKK93 pKa = 10.38SDD95 pKa = 3.68GLINDD100 pKa = 3.42NNYY103 pKa = 10.47AQTLAALFEE112 pKa = 4.7TKK114 pKa = 10.21GFPKK118 pKa = 10.62GSVNFNTYY126 pKa = 8.17TKK128 pKa = 10.27ISDD131 pKa = 4.65FINSIEE137 pKa = 4.2MTCSYY142 pKa = 10.65NIVIIPEE149 pKa = 3.94TLANSEE155 pKa = 4.34TIEE158 pKa = 4.51QVAEE162 pKa = 3.75WVLNVWKK169 pKa = 9.84CDD171 pKa = 3.2NMNVDD176 pKa = 3.42IYY178 pKa = 10.87TYY180 pKa = 10.33EE181 pKa = 3.99QVGKK185 pKa = 10.47DD186 pKa = 3.77NFWAAIGEE194 pKa = 4.39DD195 pKa = 4.29CDD197 pKa = 4.69VAVCPLDD204 pKa = 3.33TTMNGIGCTPASTEE218 pKa = 4.15TYY220 pKa = 9.71EE221 pKa = 4.26VLSNDD226 pKa = 3.46TQLALIDD233 pKa = 3.65VVDD236 pKa = 4.03NVKK239 pKa = 10.26HH240 pKa = 6.8RR241 pKa = 11.84IQLNQVTCKK250 pKa = 10.36LRR252 pKa = 11.84NCVKK256 pKa = 10.87GEE258 pKa = 3.72ARR260 pKa = 11.84LNTAIVRR267 pKa = 11.84ISNSSSFDD275 pKa = 3.44NSLSPLNNGQKK286 pKa = 8.7TRR288 pKa = 11.84SFKK291 pKa = 10.88INAKK295 pKa = 8.92KK296 pKa = 8.19WWKK299 pKa = 9.97IFYY302 pKa = 9.64TIIDD306 pKa = 4.23YY307 pKa = 10.94INTFIQSMTPRR318 pKa = 11.84HH319 pKa = 5.35RR320 pKa = 11.84AIYY323 pKa = 9.33PEE325 pKa = 3.79GWMLRR330 pKa = 11.84YY331 pKa = 9.45AA332 pKa = 4.23
MM1 pKa = 7.33VCTTLYY7 pKa = 8.57TVCVVLCILLMYY19 pKa = 10.38ILLFRR24 pKa = 11.84KK25 pKa = 9.74VIHH28 pKa = 6.25LLIDD32 pKa = 3.23MSLIAFVISSCIGLSNAQFFANDD55 pKa = 3.36MLYY58 pKa = 10.69DD59 pKa = 3.91GNVEE63 pKa = 4.23GVINTTNIFNVEE75 pKa = 4.08SLCIYY80 pKa = 9.69FPNSAVGQPGPGKK93 pKa = 10.38SDD95 pKa = 3.68GLINDD100 pKa = 3.42NNYY103 pKa = 10.47AQTLAALFEE112 pKa = 4.7TKK114 pKa = 10.21GFPKK118 pKa = 10.62GSVNFNTYY126 pKa = 8.17TKK128 pKa = 10.27ISDD131 pKa = 4.65FINSIEE137 pKa = 4.2MTCSYY142 pKa = 10.65NIVIIPEE149 pKa = 3.94TLANSEE155 pKa = 4.34TIEE158 pKa = 4.51QVAEE162 pKa = 3.75WVLNVWKK169 pKa = 9.84CDD171 pKa = 3.2NMNVDD176 pKa = 3.42IYY178 pKa = 10.87TYY180 pKa = 10.33EE181 pKa = 3.99QVGKK185 pKa = 10.47DD186 pKa = 3.77NFWAAIGEE194 pKa = 4.39DD195 pKa = 4.29CDD197 pKa = 4.69VAVCPLDD204 pKa = 3.33TTMNGIGCTPASTEE218 pKa = 4.15TYY220 pKa = 9.71EE221 pKa = 4.26VLSNDD226 pKa = 3.46TQLALIDD233 pKa = 3.65VVDD236 pKa = 4.03NVKK239 pKa = 10.26HH240 pKa = 6.8RR241 pKa = 11.84IQLNQVTCKK250 pKa = 10.36LRR252 pKa = 11.84NCVKK256 pKa = 10.87GEE258 pKa = 3.72ARR260 pKa = 11.84LNTAIVRR267 pKa = 11.84ISNSSSFDD275 pKa = 3.44NSLSPLNNGQKK286 pKa = 8.7TRR288 pKa = 11.84SFKK291 pKa = 10.88INAKK295 pKa = 8.92KK296 pKa = 8.19WWKK299 pKa = 9.97IFYY302 pKa = 9.64TIIDD306 pKa = 4.23YY307 pKa = 10.94INTFIQSMTPRR318 pKa = 11.84HH319 pKa = 5.35RR320 pKa = 11.84AIYY323 pKa = 9.33PEE325 pKa = 3.79GWMLRR330 pKa = 11.84YY331 pKa = 9.45AA332 pKa = 4.23
Molecular weight: 37.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A060NE87|A0A060NE87_9REOV RNA-directed RNA polymerase OS=Bovine rotavirus C OX=31588 GN=VP1 PE=3 SV=1
MM1 pKa = 7.5EE2 pKa = 6.6FINQTFFSEE11 pKa = 4.33SSEE14 pKa = 4.1SKK16 pKa = 9.93MDD18 pKa = 3.76VIPYY22 pKa = 10.04VLGIVLALTNGNRR35 pKa = 11.84VLKK38 pKa = 10.1IINFIITLFKK48 pKa = 11.09KK49 pKa = 9.65MVIAINVAISKK60 pKa = 8.74WKK62 pKa = 10.51RR63 pKa = 11.84EE64 pKa = 4.07NNDD67 pKa = 3.07VKK69 pKa = 11.18HH70 pKa = 5.05EE71 pKa = 4.19TKK73 pKa = 10.37DD74 pKa = 2.92IHH76 pKa = 8.3KK77 pKa = 9.91EE78 pKa = 3.81VEE80 pKa = 4.0EE81 pKa = 4.22VMTQIRR87 pKa = 11.84EE88 pKa = 4.04MRR90 pKa = 11.84IHH92 pKa = 6.57LTALFNSIHH101 pKa = 7.55DD102 pKa = 5.21DD103 pKa = 3.4NTKK106 pKa = 9.4WRR108 pKa = 11.84MSEE111 pKa = 3.9SIRR114 pKa = 11.84RR115 pKa = 11.84EE116 pKa = 3.92KK117 pKa = 10.18KK118 pKa = 10.33NEE120 pKa = 3.8MKK122 pKa = 10.69AHH124 pKa = 6.47AGVTTSKK131 pKa = 10.26QQLNNTSGLEE141 pKa = 3.99MEE143 pKa = 4.59VCLL146 pKa = 5.46
MM1 pKa = 7.5EE2 pKa = 6.6FINQTFFSEE11 pKa = 4.33SSEE14 pKa = 4.1SKK16 pKa = 9.93MDD18 pKa = 3.76VIPYY22 pKa = 10.04VLGIVLALTNGNRR35 pKa = 11.84VLKK38 pKa = 10.1IINFIITLFKK48 pKa = 11.09KK49 pKa = 9.65MVIAINVAISKK60 pKa = 8.74WKK62 pKa = 10.51RR63 pKa = 11.84EE64 pKa = 4.07NNDD67 pKa = 3.07VKK69 pKa = 11.18HH70 pKa = 5.05EE71 pKa = 4.19TKK73 pKa = 10.37DD74 pKa = 2.92IHH76 pKa = 8.3KK77 pKa = 9.91EE78 pKa = 3.81VEE80 pKa = 4.0EE81 pKa = 4.22VMTQIRR87 pKa = 11.84EE88 pKa = 4.04MRR90 pKa = 11.84IHH92 pKa = 6.57LTALFNSIHH101 pKa = 7.55DD102 pKa = 5.21DD103 pKa = 3.4NTKK106 pKa = 9.4WRR108 pKa = 11.84MSEE111 pKa = 3.9SIRR114 pKa = 11.84RR115 pKa = 11.84EE116 pKa = 3.92KK117 pKa = 10.18KK118 pKa = 10.33NEE120 pKa = 3.8MKK122 pKa = 10.69AHH124 pKa = 6.47AGVTTSKK131 pKa = 10.26QQLNNTSGLEE141 pKa = 3.99MEE143 pKa = 4.59VCLL146 pKa = 5.46
Molecular weight: 16.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5590 |
146 |
1090 |
508.2 |
58.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.136 ± 0.233 | 1.27 ± 0.354 |
5.975 ± 0.306 | 5.689 ± 0.335 |
4.436 ± 0.26 | 3.971 ± 0.414 |
1.807 ± 0.2 | 7.639 ± 0.295 |
6.655 ± 0.565 | 8.014 ± 0.473 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.47 ± 0.145 | 6.941 ± 0.346 |
2.916 ± 0.263 | 3.184 ± 0.415 |
4.741 ± 0.223 | 7.782 ± 0.512 |
6.369 ± 0.422 | 6.887 ± 0.379 |
1.127 ± 0.208 | 4.991 ± 0.505 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
